Abstract
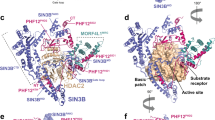
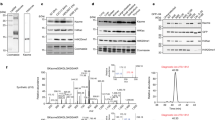
Histone acetylation plays an important role in transcriptional activation. Histones are also modified by chemically diverse acylations that are frequently deposited by p300, a transcriptional coactivator that uses a number of different acyl-CoA cofactors. Here we report that while p300 is a robust acetylase, its activity gets weaker with increasing acyl-CoA chain length. Crystal structures of p300 in complex with propionyl-, crotonyl-, or butyryl-CoA show that the aliphatic portions of these cofactors are bound in the lysine substrate-binding tunnel in a conformation that is incompatible with substrate transfer. Lysine substrate binding is predicted to remodel the acyl-CoA ligands into a conformation compatible with acyl-chain transfer. This remodeling requires that the aliphatic portion of acyl-CoA be accommodated in a hydrophobic pocket in the enzymes active site. The size of the pocket and its aliphatic nature exclude long-chain and charged acyl-CoA variants, presumably explaining the cofactor preference for p300.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Janke, R., Dodson, A.E. & Rine, J. Metabolism and epigenetics. Annu. Rev. Cell Dev. Biol. 31, 473–496 (2015).
Roth, S.Y., Denu, J.M. & Allis, C.D. Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120 (2001).
Dhalluin, C. et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496 (1999).
Strahl, B.D. & Allis, C.D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Chen, Y. et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819 (2007).
Tan, M. et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
Yang, Y.Y., Grammel, M. & Hang, H.C. Identification of lysine acetyltransferase p300 substrates using 4-pentynoyl-coenzyme A and bioorthogonal proteomics. Bioorg. Med. Chem. Lett. 21, 4976–4979 (2011).
Xie, Z. et al. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11, 100–107 (2012).
Dai, L. et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 10, 365–370 (2014).
Tan, M. et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19, 605–617 (2014).
Zhang, Z. et al. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 (2011).
Peng, C. et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell Proteomics 10 http://dx.doi.org/10.1074/mcp.M111.012658 (2011).
Moellering, R.E. & Cravatt, B.F. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science 341, 549–553 (2013).
Xie, Z. et al. Metabolic regulation of gene expression by histone lysine β-hydroxybutyrylation. Mol. Cell 62, 194–206 (2016).
Huang, H., Lin, S., Garcia, B.A. & Zhao, Y. Quantitative proteomic analysis of histone modifications. Chem. Rev. 115, 2376–2418 (2015).
Cai, L., Sutter, B.M., Li, B. & Tu, B.P. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437 (2011).
Lin, H., Su, X. & He, B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem. Biol. 7, 947–960 (2012).
Flynn, E.M. et al. A subset of human bromodomains recognizes butyryllysine and crotonyllysine histone peptide modifications. Structure 23, 1801–1814 (2015).
Goudarzi, A. et al. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Mol. Cell 62, 169–180 (2016).
Andrews, F.H. et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nat. Chem. Biol. 12, 396–398 (2016).
Li, Y. et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell 62, 181–193 (2016).
Du, J. et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 (2011).
Park, J. et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930 (2013).
Cheng, Z. et al. Molecular characterization of propionyllysines in non-histone proteins. Mol. Cell. Proteomics 8, 45–52 (2009).
Feldman, J.L., Baeza, J. & Denu, J.M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 288, 31350–31356 (2013).
Bao, X. et al. Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach. eLife 3 (2014).
Garrity, J., Gardner, J.G., Hawse, W., Wolberger, C. & Escalante-Semerena, J.C. N-lysine propionylation controls the activity of propionyl-CoA synthetase. J. Biol. Chem. 282, 30239–30245 (2007).
Smith, B.C. & Denu, J.M. Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. J. Biol. Chem. 282, 37256–37265 (2007).
Jiang, H. et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 (2013).
Yuan, H. & Marmorstein, R. Histone acetyltransferases: rising ancient counterparts to protein kinases. Biopolymers 99, 98–111 (2013).
Allis, C.D. et al. New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636 (2007).
Sabari, B.R. et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58, 203–215 (2015).
Hu, A., Britton, L.M. & Garcia, B.A. Investigating the specificity of histone acetyltransferase activity for producing rare modifications on histones using mass spectrometry. The 62nd Annual American Society for Mass Spectrometry Conference on Mass Spectrometry and Allied Topics (Baltimore, MD, 2014).
Berndsen, C.E., Albaugh, B.N., Tan, S. & Denu, J.M. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry 46, 623–629 (2007).
Leemhuis, H., Packman, L.C., Nightingale, K.P. & Hollfelder, F. The human histone acetyltransferase P/CAF is a promiscuous histone propionyltransferase. ChemBioChem 9, 499–503 (2008).
Ringel, A.E. & Wolberger, C. Structural basis for acyl-group discrimination by human Gcn5L2. Acta Crystallogr. D Struct. Biol. 72, 841–848 (2016).
Thompson, P.R. et al. Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 11, 308–315 (2004).
Liu, X. et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451, 846–850 (2008).
Delvecchio, M., Gaucher, J., Aguilar-Gurrieri, C., Ortega, E. & Panne, D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat. Struct. Mol. Biol. 20, 1040–1046 (2013).
Maksimoska, J., Segura-Peña, D., Cole, P.A. & Marmorstein, R. Structure of the p300 histone acetyltransferase bound to acetyl-coenzyme A and its analogues. Biochemistry 53, 3415–3422 (2014).
Thompson, P.R., Kurooka, H., Nakatani, Y. & Cole, P.A. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. J. Biol. Chem. 276, 33721–33729 (2001).
Gould, T.A., Schweizer, H.P. & Churchill, M.E. Structure of the Pseudomonas aeruginosa acyl-homoserinelactone synthase LasI. Mol. Microbiol. 53, 1135–1146 (2004).
Watson, W.T., Minogue, T.D., Val, D.L., von Bodman, S.B. & Churchill, M.E. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol. Cell 9, 685–694 (2002).
Carvalho, P.C., Hewel, J., Barbosa, V.C. & Yates, J.R. III. Identifying differences in protein expression levels by spectral counting and feature selection. Genet. Mol. Res. 7, 342–356 (2008).
Karukurichi, K.R. & Cole, P.A. Probing the reaction coordinate of the p300/CBP histone acetyltransferase with bisubstrate analogs. Bioorg. Chem. 39, 42–47 (2011).
Zhang, X. et al. Catalytic mechanism of histone acetyltransferase p300: from the proton transfer to acetylation reaction. J. Phys. Chem. B 118, 2009–2019 (2014).
Rousseaux, S. & Khochbin, S. Histone acylation beyond acetylation: aerra incognita in chromatin biology. Cell J. 17, 1–6 (2015).
Yang, C. et al. Labeling lysine acetyltransferase substrates with engineered enzymes and functionalized cofactor surrogates. J. Am. Chem. Soc. 135, 7791–7794 (2013).
King, M.T. & Reiss, P.D. Separation and measurement of short-chain coenzyme-A compounds in rat liver by reversed-phase high-performance liquid chromatography. Anal. Biochem. 146, 173–179 (1985).
Hosokawa, Y., Shimomura, Y., Harris, R.A. & Ozawa, T. Determination of short-chain acyl-coenzyme A esters by high-performance liquid chromatography. Anal. Biochem. 153, 45–49 (1986).
Luger, K., Rechsteiner, T.J. & Richmond, T.J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 (1999).
Acknowledgements
We thank the staff of ESRF and EMBL-Grenoble for assistance and support in using beamlines ID29, 23-1 and 23-2 and the HTX facility. We thank L. Signor (IBS, Grenoble) and P. Phapale (EMBL) for MS analysis. This work used the platforms of the Grenoble Instruct centre (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB). Z.K. was supported by a fellowship from the EMBL Interdisciplinary Postdoc Programme under Marie Skłodowska-Curie actions Cofund (grant agreement number 291772). This work was supported by the ANR Grant Episperm3 (ANR-15-CE12- 0005-02) to S. Khochbin and D.P. and the Worldwide Cancer Research foundation (grant #16-0280) to D.P. Work in S. Khochbin's laboratory is supported by INCa, Fondation pour la Recherche Médicale and Fondation ARC. Work in Y.Z.'s laboratory is supported by NIH grants GM105933, DK107868 and GM115961.
Author information
Authors and Affiliations
Contributions
Z.K. performed the structure determination of p300 under supervision of J.A.M. and D.P. E.O. and Z.K. performed biochemical assays. E.O. prepared the constructs and performed protein expression and purification. A.G. performed the IP–HAT experiments under the supervision of S. Khochbin. H.H. and S. Kim performed MS experiments under the supervision of Y.Z. D.P. designed and coordinated the project, advised and assisted on all aspects of the project and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Table 1 and Supplementary Figures 1–7. (PDF 1196 kb)
Supplementary Note
Annotated MS/MS spectra of for histone KAc from experiments shown in Figure 5. (PDF 2579 kb)
Rights and permissions
About this article
Cite this article
Kaczmarska, Z., Ortega, E., Goudarzi, A. et al. Structure of p300 in complex with acyl-CoA variants. Nat Chem Biol 13, 21–29 (2017). https://doi.org/10.1038/nchembio.2217
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2217