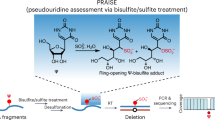
Abstract
Pseudouridine (Ψ) is the most abundant post-transcriptional RNA modification, yet little is known about its prevalence, mechanism and function in mRNA. Here, we performed quantitative MS analysis and show that Ψ is much more prevalent (Ψ/U ratio ∼0.2–0.6%) in mammalian mRNA than previously believed. We developed N3-CMC–enriched pseudouridine sequencing (CeU-Seq), a selective chemical labeling and pulldown method, to identify 2,084 Ψ sites within 1,929 human transcripts, of which four (in ribosomal RNA and EEF1A1 mRNA) are biochemically verified. We show that hPUS1, a known Ψ synthase, acts on human mRNA; under stress, CeU-Seq demonstrates inducible and stress-specific mRNA pseudouridylation. Applying CeU-Seq to the mouse transcriptome revealed conserved and tissue-specific pseudouridylation. Collectively, our approaches allow comprehensive analysis of transcriptome-wide pseudouridylation and provide tools for functional studies of Ψ-mediated epigenetic regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Machnicka, M.A. et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
Charette, M. & Gray, M.W. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49, 341–351 (2000).
Ge, J. & Yu, Y.T. RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 38, 210–218 (2013).
Jack, K. et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 44, 660–666 (2011).
Yu, A.T., Ge, J. & Yu, Y.T. Pseudouridines in spliceosomal snRNAs. Protein Cell 2, 712–725 (2011).
Kierzek, E. et al. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 42, 3492–3501 (2014).
Kiss, T., Fayet-Lebaron, E. & Jady, B.E. Box H/ACA small ribonucleoproteins. Mol. Cell 37, 597–606 (2010).
Hamma, T. & Ferre-D′Amare, A.R. Pseudouridine synthases. Chem. Biol. 13, 1125–1135 (2006).
Heiss, N.S. et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 19, 32–38 (1998).
Bykhovskaya, Y., Casas, K., Mengesha, E., Inbal, A. & Fischel-Ghodsian, N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am. J. Hum. Genet. 74, 1303–1308 (2004).
Wu, G., Xiao, M., Yang, C. & Yu, Y.T. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 30, 79–89 (2011).
Courtes, F.C. et al. 28S rRNA is inducibly pseudouridylated by the mTOR pathway translational control in CHO cell cultures. J. Biotechnol. 174, 16–21 (2014).
Basak, A. & Query, C.C. A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep. 8, 966–973 (2014).
Cavaillé, J. et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 97, 14311–14316 (2000).
Hüttenhofer, A., Brosius, J. & Bachellerie, J.P. RNomics: identification and function of small, non-messenger RNAs. Curr. Opin. Chem. Biol. 6, 835–843 (2002).
Karijolich, J. & Yu, Y.-T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398 (2011).
Fernández, I.S. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature 500, 107–110 (2013).
Ho, N.W. & Gilham, P.T. The reversible chemical modification of uracil, thymine, and guanine nucleotides and the modification of the action of ribonuclease on ribonucleic acid. Biochemistry 6, 3632–3639 (1967).
Bakin, A. & Ofengand, J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32, 9754–9762 (1993).
Bakin, A.V. & Ofengand, J. Mapping of pseudouridine residues in RNA to nucleotide resolution. Methods Mol. Biol. 77, 297–309 (1998).
Carlile, T.M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Lovejoy, A.F., Riordan, D.P. & Brown, P.O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE 9, e110799 (2014).
Hughes, D.G. & Maden, B.E. The pseudouridine contents of the ribosomal ribonucleic acids of three vertebrate species. Numerical correspondence between pseudouridine residues and 2′-O-methyl groups is not always conserved. Biochem. J. 171, 781–786 (1978).
Maden, B.E. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 39, 241–303 (1990).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Desrosiers, R., Friderici, K. & Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 71, 3971–3975 (1974).
Dubin, D.T. & Taylor, R.H. The methylation state of poly A–containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 2, 1653–1668 (1975).
Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 (2010).
Liu, N. et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013).
Hunter, S. et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, D306–D312 (2012).
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Baltz, A.G. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012).
Zhao, X. et al. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol. Cell 15, 549–558 (2004).
Sibert, B.S. & Patton, J.R. Pseudouridine synthase 1: a site-specific synthase without strict sequence recognition requirements. Nucleic Acids Res. 40, 2107–2118 (2012).
Becker, H.F., Motorin, Y., Planta, R.J. & Grosjean, H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of Ψ55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 25, 4493–4499 (1997).
Behm-Ansmant, I. et al. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA 9, 1371–1382 (2003).
Dominissini, D. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K.D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
He, C. Grand challenge commentary: RNA epigenetics? Nat. Chem. Biol. 6, 863–865 (2010).
Fu, Y., Dominissini, D., Rechavi, G. & He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293–306 (2014).
Liu, N. et al. N6-methyladenosine–dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Tremml, G., Singer, M. & Malavarca, R. Culture of mouse embryonic stem cells. Curr. Protoc. Stem Cell Biol. 5, 1C4 (2008).
König, J. et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17, 909–915 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Ding, Y. et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700 (2014).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Acknowledgements
The authors would like to thank R. Meng and S. Huang for technical assistance and N. Liu, X. Wang and X. Liu for discussion. This work was supported by the National Basic Research Foundation of China (no. 2014CB964900) and the National Natural Science Foundation of China (nos. 21472009 and 31270838).
Author information
Authors and Affiliations
Contributions
X.L., S.M. and C.Y. conceived the project, designed the experiments and wrote the manuscript. X.L. and S.M. performed the experiments. P.Z. designed and performed the bioinformatics analysis; J.S. contributed in making figures, J.B. synthesized the N3-CMC derivative, and F.S. contributed in Ψ quantification. All authors commented on and approved the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–6, Supplementary Figures 1–11 and Supplementary Note. (PDF 2795 kb)
Rights and permissions
About this article
Cite this article
Li, X., Zhu, P., Ma, S. et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11, 592–597 (2015). https://doi.org/10.1038/nchembio.1836
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1836
This article is cited by
-
BID-seq for transcriptome-wide quantitative sequencing of mRNA pseudouridine at base resolution
Nature Protocols (2024)
-
Small RNA modifications: regulatory molecules and potential applications
Journal of Hematology & Oncology (2023)
-
RNA modification in cardiovascular disease: implications for therapeutic interventions
Signal Transduction and Targeted Therapy (2023)
-
Quantitative profiling of pseudouridylation landscape in the human transcriptome
Nature Chemical Biology (2023)
-
Bioinformatic analysis of m6A “reader” YTH family in pan-cancer as a clinical prognosis biomarker
Scientific Reports (2023)