Abstract
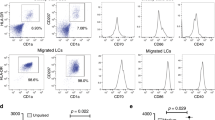
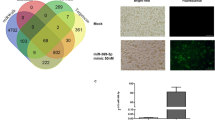
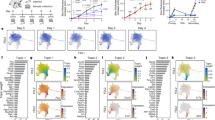
Langerhans cells (LCs) are bone marrow-derived immature skin-residential dendritic cells (DCs) with a life cycle distinct from that of other types of DCs. The mechanisms involved in LC homeostasis and immunological functions are still not clear. MicroRNAs (miRNAs) are a class of short noncoding RNAs that regulate gene expression through either translational repression or mRNA degradation. A recent study showed that specific deletion of total miRNAs in DCs affects the homeostasis and function of only LCs, but not of other types of DCs. The roles of specific individual miRNA in LC development are still lacking. The miRNA miR-17-92 class, encoding miR-17, miR-18, miR-19a, miR-19b, miR-20 and miR-92, plays a very important role in B- and T-cell development and function. Here, we first report that epidermal LCs highly express the miR-17-92 class compared with spleen naive T cells. To further characterize the role of miR-17-92 in LC development, we generated LC-specific miR-17-92 knockout and knock-in mice. Interestingly, LC-specific gain- and loss-of-function of miR-17-92 cluster did not significantly change LC homeostasis, maturation ability, antigen capture and migration to draining lymph nodes. Thus, the miR-17-92 cluster may be functionally redundant and not critically required for LC development and function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Zhou L, Park JJ, Zheng Q, Dong Z, Mi Q . MicroRNAs are key regulators controlling iNKT and regulatory T-cell development and function. Cell Mol Immunol 2011; 8: 380–387.
Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY . Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med 2008; 205: 1993–2004.
Zhou L, Seo KH, He HZ, Pacholczyk R, Meng DM, Li CG et al. Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development. Proc Natl Acad Sci USA 2009; 106: 10266–10271.
Zheng Q, Zhou L, Mi QS . MicroRNA miR-150 is involved in Valpha14 invariant NKT cell development and function. J Immunol 2012; 188: 2118–2126.
Romani N, Clausen BE, Stoitzner P . Langerhans cells and more: langerin-expressing dendritic cell subsets in the skin. Immunol Rev 2010; 234: 120–141.
Kaplan DH . In vivo function of Langerhans cells and dermal dendritic cells. Trends Immunol 2010; 31: 446–451.
Kuipers H, Schnorfeil FM, Fehling HJ, Bartels H, Brocker T . Dicer-dependent microRNAs control maturation, function and maintenance of Langerhans cells in vivo. J Immunol 2010; 185: 400–409.
Mi QS, Xu YP, Qi RQ, Shi YL, Zhou L . Lack of microRNA miR-150 reduces the capacity of epidermal Langerhans cell cross-presentation. Exp Dermatol 2012; 21: 876–877.
Mi QS, Xu YP, Wang H, Qi RQ, Dong Z, Zhou L . Deletion of microRNA miR-223 increases Langerhans cell cross-presentation. Int J Biochem Cell Biol 2013; 45: 395–400.
Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 2008; 9: 405–414.
Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008; 132: 875–886.
Kaplan DH, Jenison MC, Saeland S, Shlomchik WD, Shlomchik MJ . Epidermal langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity 2005; 23: 611–620.
Kaplan DH, Li MO, Jenison MC, Shlomchik WD, Flavell RA, Shlomchik MJ . Autocrine/paracrine TGFbeta1 is required for the development of epidermal Langerhans cells. J Exp Med 2007; 204: 2545–2552.
Kissenpfennig A, it-Yahia S, Clair-Moninot V, Stossel H, Badell E, Bordat Y et al. Disruption of the langerin/CD207 gene abolishes Birbeck granules without a marked loss of Langerhans cell function. Mol Cell Biol 2005; 25: 88–99.
Stoitzner P, Tripp CH, Eberhart A, Price KM, Jung JY, Bursch L et al. Langerhans cells cross-present antigen derived from skin. Proc Natl Acad Sci USA 2006; 103: 7783–7788.
Acknowledgements
We thank all members from our laboratories for their advice and encouragement. This work was supported by grants from the National Institutes of Health Grant 1R21AR059976 and Henry Ford Health System Start-up Grant for the Immunology Program (T71016 and T71017).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Zhou, L., Qi, RQ., Liu, M. et al. microRNA miR-17-92 cluster is highly expressed in epidermal Langerhans cells but not required for its development. Genes Immun 15, 57–61 (2014). https://doi.org/10.1038/gene.2013.61
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2013.61
Keywords
This article is cited by
-
Transcription factor and miRNA co-regulatory network reveals shared and specific regulators in the development of B cell and T cell
Scientific Reports (2015)
-
An updated review of mechanotransduction in skin disorders: transcriptional regulators, ion channels, and microRNAs
Cellular and Molecular Life Sciences (2015)