Abstract
While considered an extracellular pathogen, Pseudomonas aeruginosa has been reported to be engulfed by macrophages in cellular and animal models. However, the role of macrophages in P. aeruginosa clearance in vivo remains poorly studied. The major outer membrane porin OprF has been recently shown to be involved in P. aeruginosa fate within cultured macrophages and analysis of an oprF mutant may thus provide insights to better understand the relevance of this intramacrophage stage during infection. In the present study, we investigated for the first time the virulence of a P. aeruginosa oprF mutant in a vertebrate model that harbors functional macrophages, the zebrafish (Danio rerio) embryo, which offers powerful tools to address macrophage–pathogen interactions. We established that P. aeruginosa oprF mutant is attenuated in zebrafish embryos in a macrophage-dependent manner. Visualization and quantification of P. aeruginosa bacteria phagocytosed by macrophages after injection into closed cavities suggested that the attenuated phenotype of oprF mutant is not linked to higher macrophage recruitment nor better phagocytosis than wild-type strain. Using cultured macrophages, we showed an intramacrophage survival defect of P. aeruginosa oprF mutant, which is correlated with elevated association of bacteria with acidic compartments. Notably, treatment of embryos with bafilomycin, an inhibitor of acidification, increased the sensibility of embryos towards both wild-type and oprF mutant, and partially suppressed the attenuation of oprF mutant. Taken together, this work supports zebrafish embryo as state-of-the-art model to address in vivo the relevance of P. aeruginosa intramacrophage stage. Our results highlight the contribution of macrophages in the clearance of P. aeruginosa during acute infection and suggest that OprF protects P. aeruginosa against macrophage clearance by avoiding bacterial elimination in acidified phagosomes.
Similar content being viewed by others
Introduction
The environmental bacterium P. aeruginosa is an opportunistic human pathogen responsible for a variety of acute infections and is a major cause of mortality in chronically infected cystic fibrosis patients. Numerous reports have emphasized that the extracellular pathogen P. aeruginosa can enter host cells, resulting in a phase of intracellular residence, which can be of importance in addition to the classical extracellular infection. An intracellular stage of P. aeruginosa within cultured epithelial cells has been known for a long time1,2,3 and advanced imaging methods have allowed to track bacteria within epithelial cells4. More recently, P. aeruginosa has been also localized within cultured macrophages5,6,7. The intramacrophage fate of the bacteria has revealed vacuolar escape of P. aeruginosa and macrophage death driven by intracellular bacteria, most likely linked to cytosolic location of bacteria8. Bacterial factors involved in this intramacrophage step have been recently investigated6,8. MgtC and OprF have been uncovered as bacterial factors involved in the intramacrophage survival of P. aeruginosa6,7,9. Our work recently established that MgtC and OprF modulate the transcription of type III secretion system (T3SS) genes. T3SS, and more specifically its ExoS effector, play a main role in the intramacrophage life of P. aeruginosa, allowing internalized bacteria to escape phagosomes and promote macrophages lysis8. Consistent with the effect of OprF on T3SS genes transcription, OprF modulated the production of the T3SS PcrV cap protein and the secretion of ExoT and ExoS toxins10,11.
OprF is a major outer membrane porin involved in maintenance of cell structure, outer membrane permeability, environmental sensing, adhesion, biofilm formation and virulence12,13. Besides regulating the secretion of T3SS effectors, OprF also modulates the production of the quorum-sensing-dependent virulence factors pyocyanin, elastase, lectin PA-1L, and exotoxin A10. Accordingly, in the oprF mutant, production of the quorum-sensing signal molecules is reduced or delayed10,12. The virulence of oprF mutant is reduced in an invertebrate model organism, the nematode C. elegans10. Moreover, a reduction in the cytotoxicity was observed for the oprF mutant on various cultivated mammalian cell lines, including epithelial cells and macrophages7,10.
The role of macrophages in vivo in internalization and early clearance of P. aeruginosa remains poorly studied, as shown with the controversial results regarding the contribution of mouse lung alveolar macrophages in P. aeruginosa infection14,15. In the present study, we explored the behavior of an oprF mutant in connexion with intramacrophage survival during P. aeruginosa acute infection in a non-mammalian vertebrate animal model. Zebrafish (Danio rerio) embryo is a model of choice for studying interactions between pathogens and host innate immune system, due to its optical transparency that allows the analysis of infections in real time using fluorescent microorganisms16,17. Moreover, zebrafish innate immune system is closely related to the one of mammals and transgenic reporter lines are available to visualize innate immune cells16,17. This model has therefore been highlighted as a suitable model to address the interaction between bacteria and macrophages18,19,20. P. aeruginosa has been shown to be phagocytosed by macrophages upon acute infection in zebrafish embryos21,22, thus allowing to study in vivo the bacterial factors involved in P. aeruginosa survival into macrophages and evaluate their importance in the outcome of infection. In the present study, we first investigated the behavior of oprF mutant strain in vivo upon infection of zebrafish embryo, before moving to further analysis with cultured macrophages. Our results highlight the contribution of macrophages in the clearance of P. aeruginosa during acute infection and support a role of OprF in the ability of P. aeruginosa to avoid localization in acidified phagosomes and resist elimination by macrophages.
Results
In the Danio rerio infection model, OprF is important for P. aeruginosa virulence in a macrophage-dependent manner
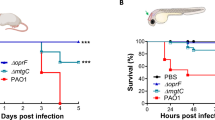
We evaluated the role of OprF in P. aeruginosa virulence in the zebrafish (Danio rerio) embryo model. Zebrafish embryo is a model of choice, which has been used for various intracellular and extracellular bacterial pathogens, to investigate the contribution of innate immune cells during infection19,20. In a previous study, we have introduced a plasmid constitutively producing GFP8 in an oprF mutant in the PAO1 background11. Here, the same mutant and the isogenic wild-type strain were injected intravenously in the caudal vein of embryos at 50 h post-fertilization (Fig. 1A). The survival curves of infected embryos indicate that OprF is a critical virulence determinant in this model since the virulence of oprF mutant is significantly attenuated as compared to the one of wild-type PAO1 strain (Fig. 1B, left graph). In addition, fluorescence microscopy of infected embryos (20 h post-infection) showed a lower bacterial burden with oprF mutant than wild-type strain (Fig. 1C, quantification in Fig. 1D). A similar result was observed when bacterial burden was assessed by colony forming unit (CFU) counting (Fig. S1).
The P. aeruginosa ΔoprF mutant is attenuated in zebrafish embryos in a macrophage-dependent manner. (A) Diagram of 50 h post-fertilization (hpf) zebrafish embryo showing the caudal vein injection site (green arrow). (B) Survival curves of embryos infected with PAO1 wild-type stain or PAO1 ΔoprF mutant. Non infected embryos (NI) were used as control. Embryos were either treated with LipoPBS (left panel) or with LipoCld (right panel) to deplete macrophages. Approximately 3500 CFU P. aeruginosa were microinjected into the caudal vein (n = 50–70 for infected embryos and n = 30 for NI embryos, pool of three independent experiments). Results are expressed as the percentage of surviving embryos at different times post-infection. (C) Representative fluorescence images of embryos infected by either wild-type or ΔoprF P. aeruginosa (at 20 hpi) in the context of LipoPBS or LipoCld treatment. The efficiency of macrophages depletion is shown in the upper panels with the visualization of red macrophages. Scale bar 400 µm. (D) Quantification of bacterial loads (fluorescence pixel counts) from two independent experiments. Each symbol represents individual embryo (n = 18–20) and horizontal lines indicate the median values. Statistical significance was determined by log-rank test (B) or one-tailed Mann–Whitney’s test (D).
To evaluate the contribution of macrophages in the attenuated phenotype of oprF mutant, we took advantage on the fact that macrophages can be depleted from zebrafish embryos with a validated method that uses liposome-encapsulated clodronate (LipoCld)23,24. Macrophage-depleted zebrafish embryos have been shown to be highly sensitive to P. aeruginosa infection6,21. We have carried out experiments with tg(mfap4:mCherry-F) embryos where macrophages are visualized as red cells25, which allows to check macrophage depletion upon LipoCld injection (Fig. 1C, upper panels). Interestingly, the survival curve of macrophage-depleted embryos showed a dramatic increase in the virulence of oprF mutant strain, since all embryos were killed, with a slight delay comparatively to the wild-type strain (Fig. 1B, right graph). Thus, macrophage depletion largely attenuates the difference of survival between wild-type and mutant strain. In agreement with this finding, bacterial burden was elevated and not significantly different for both wild-type and mutant strains in the context of macrophage depletion (Fig. 1C,D).
Taken together, these results indicate that OprF plays a crucial role during acute infection of P. aeruginosa in the zebrafish embryo model and that oprF mutant strain is better cleared than wild-type strain in a macrophage-dependent manner.
Visualisation of macrophages in infected zebrafish embryos indicates no major difference in macrophage recruitment or phagocytosis efficiency between wild-type P. aeruginosa and oprF mutant
Confocal microscopy after local injection into closed cavities, such as hindbrain ventricle (HBV) or muscle allows to visualize recruited macrophages and bacteria phagocytosed by macrophages close to the site of injection26. Upon HBV infection, wild-type and ΔoprF bacteria can be visualized within macrophages 2 h after infection (Fig. S2). A similar finding is observed upon muscle injection. To visualize macrophage recruitment and phagocytosed bacteria in real-time, a time lapse experiment was performed during 3 h. Injection in the muscle was preferred to HBV injection because of easier positioning of embryos and lower thickness that facilitates z-stacks analysis (Fig. 2A). The recruitment of macrophages that phagocytose P. aeruginosa can be clearly observed, both for clustered bacteria and isolated bacteria, as shown for ΔoprF bacteria (Fig. 2B). The number of recruited macrophages and the clearance of oprF mutant by macrophages were then compared to the one of wild-type strain by performing quantification on images from time 1.5 h and 4.5 h on 16 to 18 embryos for each strain (Fig. 2C). The number of recruited macrophages is similar for both strains at time 1.5 h and slightly, but significantly, higher for wild-type strain at 4.5 h (Fig. 2D). Similarly, the number of infected macrophages appeared similar for both strains at time 1.5 h and slightly, but significantly, higher for wild-type strain at 4.5 h (Fig. 2E). In contrast, a slight, but significant, reduction for bacteria counting was observed for the mutant strain comparatively to wild-type strain at 4.5 h (Fig. 2F), indicative of a better clearance of bacteria. This is correlated with a tendency of increased green Fluorescent Pixel Counts (FPC) for wild-type strain and decreased FCP for oprF mutant at 4.5 h relatively to FPC at 1.5 h (Fig. 2F, right panel).
Real-time visualization and quantification of phagocytosis of P. aeruginosa after local injection into muscle of wild-type and ΔoprF strains. (A) Illustration of zebrafish larva with the muscle (in red) injection site (green arrow). (B) Confocal time-lapse images of tg(mfap4:mCherry-F) larva (red macrophages) infected with ΔoprF P. aeruginosa (green) by injection in the muscle. The same area was imaged every hour from 1.5 hpi. White arrows depict GFP-expressing P. aeruginosa that will be taken up by macrophages, which are recruited at the bacterial location. Maximum intensity projection of 96 sections every 1 µm, scale bar 50 µm. (C) Confocal time-lapse images of tg(mfap4:mCherry-F) larvae (red macrophages) infected with wild-type or ΔoprF P. aeruginosa (green) by injection in the muscle. The left panel is at 1.5 hpi and the right panel a 4.5 hpi. White arrows depict GFP-expressing P. aeruginosa phagocytosed by macrophages. Maximum intensity projection of 96 sections every 1 µm, scale bar 50 µm. (D) Quantification of recruited macrophages at 1.5 and 4.5 hpi, from three independent experiments. Each symbol represents individual embryo (16 to 18 embryos for each strain) and horizontal lines indicate the mean values ± SEM. Statistical significance was determined by two-tailed t test, ns—not significant, *p < 0.05. (E) Quantification of infected macrophages, from three independent experiments. Mean ± SEM, two-tailed Mann–Whitney’s test, ns—not significant, *p < 0.05. (F) Quantification of bacterial loads (fluorescence pixel counts) from three independent experiments, presented as total value (left) or percentage of the FPC value at first time point (1.5 hpi), counted for each embryo. Mean ± SEM, two-tailed Mann–Whitney’s test, ns—not significant, *p < 0.05.
Cumulatively, our results revealed that confocal microscopy after local injection into hindbrain ventricle or muscle is a highly suitable approach to visualize and quantify in real-time P. aeruginosa phagocytosis by recruited macrophages. The better clearance of the oprF mutant does not seem to be linked to an increase in macrophage recruitment or phagocytosis efficiency. To dissect intramacrophage mechanisms underlying oprF mutant clearance, we carried out experiments on cultured macrophages.
P. aeruginosa oprF mutant exhibits reduced resistance to cultured macrophages, which is correlated with elevated association of bacteria with acidic compartments
An otopathogenic P. aeruginosa oprF mutant strain was found to be more sensitive to macrophage killing than the wild-type strain upon infection of mouse bone marrow macrophages7. In a previous study, we have used the oprF mutant in the PAO1 background to address the cytotoxicity driven by phagocytosed bacteria in macrophages8. Here, we quantified intracellular bacteria after phagocytosis of P. aeruginosa strains. J774 macrophages were infected with wild-type PAO1 and oprF mutant strains expressing constitutively GFP grown exponentially in LB medium (Multiplicity of infection or MOI = 10). After 25 min of phagocytosis, several washes were performed to remove adherent bacteria and gentamicin was added to kill extracellular bacteria. Microscopic observation of infected macrophages was done after 20 min or 2.5 h of gentamicin treatment (Fig. 3A). The number of bacteria per macrophage was quantified and classified in three groups (Fig. 3B). Wild-type and mutant strains behaved similarly at the early time point. At the latest time, macrophages infected with oprF mutant were found to harbor less bacteria than macrophages infected with wild-type strain. This result supports a role for OprF to limit the elimination of P. aeruginosa PAO1 by macrophages.
Visualization and quantification of intracellular P. aeruginosa within macrophages. GFP-expressing PAO1 WT and ∆oprF strains were used for infecting J774 macrophages. Cells were fixed after phagocytosis and 20 min (T0) or 2.5 h (T3h) treatment with gentamycin and imaged with fluorescent microscope. DAPI was used to stain the nucleus. (A) Visualization of intracellular bacteria. The merged image shows Differential Interference Contrast (DIC), nucleus staining (blue) and bacteria expressing GFP (green). The scale bars depict 10 µm. (B) Count of the number of bacteria in infected macrophages from images obtained. The numbers of bacteria per cell were classified in three groups (1–2, 3–5 and > 6 bacteria per cell) and percentage of each class is shown. Count is done from 100 cells per condition and results are expressed as means from three independent experiments.
We further examined intracellular P. aeruginosa oprF mutant within macrophages in more detail using transmission electron microscopy (TEM). J774 macrophages infected with wild-type PAO1 strain or oprF mutant were subjected to fixation at different time points after phagocytosis. At early time point (50 min post phagocytosis) intracellular wild-type P. aeruginosa are essentially observed in membrane bound vacuoles (Fig. 4A–C). For the oprF mutant, bacteria were frequently observed in vacuoles partially or totally filled with heterogeneous electron dense material, suggesting that the vacuoles had fused with lysosomes (Fig. 4D–F). At time 2.5 h post phagocytosis, intracellular bacteria were rarely identifiable for the oprF mutant, suggesting that most of them have been destroyed, while for the wild-type strain, numerous bacteria were observed in clear vacuoles or in the cytoplasm with no surrounding membrane as previously described8. In the course of TEM analysis of J774 macrophages infected by wild-type PAO1 strain, we noticed the presence of vesicles in the vacuolar space along the phagosomal membrane (Fig. 4C,F). These vesicles seemed issued from bacteria and were often lying along the phagosomal membrane (Fig. S3, A-B). Because OprF is a major component of outer membrane vesicles (OMVs)27, we investigated whether such bacterial vesicles were also present in the oprF mutant. Vesicles were clearly produced from oprF mutant as well and found associated with the phagosomal membrane (Fig. S3, C-D).
Transmission electron micrographs (TEM) of P. aeruginosa within macrophages. J774 macrophages were infected with P. aeruginosa wild-type strain or oprF mutant for 50 min after phagocytosis and subjected to TEM to visualize intracellular bacteria. For the wild-type strain, most of bacteria were found inside membrane bound vacuoles (A–C) whereas oprF mutant was mostly found in phagolysosomes (D–F). (B) and (C) are showing details of the cell shown in (A) (white rectangles) at higher magnification, (E) and (F) show details of the cell shown in (D) (white rectangles).
To quantify the association between bacteria and acidic compartments such as phagolysosomes, we examined the association between fluorescent PAO1 wild-type and oprF mutant bacteria and the LysoTracker probe during infection using fixed macrophages. Bacteria colocalizing with LysoTracker could be visualized (Fig. 5A) and quantified (Fig. 5B). A significantly higher percentage of bacteria colocalizing with LysoTracker red marker was observed in macrophages infected with oprF mutant (~ 75%) comparatively to wild-type strain (~ 45%). This increased localization of oprF mutant in acidified compartments corroborates the TEM observation of a preferential localization of ΔoprF bacteria in phagolysosomes.
Colocalization of P. aeruginosa with a probe that labels acidic compartments. J774 macrophages were infected with PAO1 and ∆oprF strains expressing GFP. After 2.5 h of gentamicin treatment, infected J774 cells were incubated with Lysotracker for 10 min, a red fluorescent weak base that accumulates in acidic compartments. Cells were then fixed and imaged with fluorescence microscope. (A) The image shows individual panels for Differential Interference Contrast (DIC), lysosomal compartment (red), bacteria expressing GFP (green), the nucleus (blue) and merged image of all channels. White arrows show colocalization of bacteria with lysotracker while yellow arrows show non-colocalization. Scale bar is equivalent to 5 µm. (B) Values indicate the percentage of bacteria that colocalized with Lysotracker red (white arrows). Data are the mean (± SEM) of 3 independent experiments, with a minimum of 100 bacteria counted per experiment for each sample. *, p ≤ 0.05 (Two-tailed unpaired t test).
Taken together, this results indicate an intramacrophage survival defect of oprF mutant, which is associated with a preferential localization within acidified compartments.
Contribution of phagosomal acidification to the outcome of infection in vivo
Because oprF mutant attenuation in zebrafish is dependent on the presence of macrophages and because the oprF mutant colocalizes more frequently than wild-type strain with acidic compartments in infected macrophages, we hypothesized that decreasing phagosomal acidification in zebrafish embryos may alleviate the attenuation of oprF mutant. The acidification of the phagosome is dependent on the activity of the host vacuolar ATPase, which can be specifically inhibited by the inhibitor bafilomycin A128. Bafilomycin was added to the larvae bath water as reported earlier29 to test the implication of macrophage phagosomal acidification as a microbicidal mechanism responsible for the attenuated phenotype of oprF mutant in zebrafish larvae. The survival of embryos infected with wild-type strain and oprF mutant was reduced in the presence of bafilomycin, supporting the idea that phagosomal acidification is important for host defense (Fig. 6A,B). We reasoned that if increased elimination by acidified phagosomes contributes to oprF mutant attenuation, then bafilomycin treatment should suppress this attenuated phenotype. This is partially the case, since the virulence of oprF mutant is greatly increased in the presence of bafilomycin (Fig. 6B). The bacterial burden was evaluated by microscopy analysis and quantification of the FPC (Fig. 6C). Bacterial fluorescence counts are significantly lower for oprF mutant than wild-type strain upon HBV injection, which is correlated with lower CFU (Fig. S4), and is consistent with results upon caudal vein injection (Fig. 1D). In contrast, the number of fluorescent pixels for the oprF mutant was strongly increased in the presence of bafilomycin, becoming significantly higher than the one of wild-type strain (Fig. 6C,D). These data support the hypothesis that acidification contributes to limit the growth of oprF mutant in non-treated embryos.
Effect of bafilomycin on the virulence of wild-type and ΔoprF mutant in zebrafish embryos. Embryos were infected with either wild-type or ΔoprF P. aeruginosa (2000–2500 CFU) in the hindbrain ventricle at 2 dpf or not infected (NI) (n = 60–70 for infected embryos and n = 45 for NI embryos, pool of three independent experiments). Survival curves of embryos treated with 50 nM bafilomycin (Bafil) (B) or treated with DMSO as control (A). Statistical significance was determined by log-rank test. Imaging (C) and quantification (D) of bacterial loads (fluorescence pixel counts) representative of two independent experiments at 20 hpi (n = 11–14). Each symbol represents individual embryo and horizontal lines indicate the median values. Statistical significance was determined by one-tailed Mann–Whitney’s test.
Cumulatively, results carried out with cellular and animal models indicate that OprF acts by protecting P. aeruginosa against macrophages clearance during acute infection, in part by avoiding acidified phagosomes.
Discussion
Recent studies support the ability of P. aeruginosa to reside, at least transiently, in macrophages and bacterial factors that play a role in the intramacrophage stage, such as OprF, are starting to be identified. However, their role towards macrophage bactericidal action remains mostly elusive. In the present study, we have taken advantage of the zebrafish embryo model, in combination with a cellular infection approach, and orpF mutant to better understand how P. aeruginosa resists to macrophage clearance during acute infection.
We have evaluated for the first time the role of OprF in P. aeruginosa virulence in a vertebrate model harbouring functional macrophages, the zebrafish embryo model. This model, which has been used for various intracellular and extracellular bacterial pathogens, is a model of choice to investigate the contribution of innate immune cells during infection20. Zebrafish embryo is increasingly considered for modeling human infections caused by bacterial pathogens, including those affecting the lungs30,31. Moreover, P. aeruginosa bacteria have been visualised within macrophages upon zebrafish embryo infection6,21,22,32,33, suggesting a relevance for this model to evaluate the role of intramacrophage step in the outcome of infection. Our results showed an attenuation of the oprF mutant in zebrafish embryos, which is dependent on the presence of macrophages, thus supporting the importance of OprF in resistance to macrophage. Injection of bacteria into closed cavities, such as hindbrain ventricle, or into muscle provides an outstanding opportunity to visualize macrophage recruitment and bacterial phagocytosis in real-time26. Images of infected embryos clearly support the importance of macrophages in P. aeruginosa clearance, with a large proportion of bacteria being engulfed by macrophages. Based on measurement of bacterial load, the oprF mutant appeared to be cleared more efficiently than wild-type strain. However, it was not associated with higher recruitment of macrophages or higher phagocytosis rate of the oprF mutant strain by macrophages in vivo. The ability of OprF to bind the complement component C3b, which tags bacteria for phagocytosis by host macrophages and neutrophils, was previously proposed to be responsible for a reduced internalization by neutrophils after opsonization of oprF-deficient bacteria compared with wild-type P. aeruginosa34. Because the internalization within macrophages does not seem to be impacted in our study, the behavior of oprF mutant strain in zebrafish embryos is unlikely linked to the complement system, possibly due to the fact that bacteria are not opsonized.
OprF has been previously involved in the intramacrophage survival otopathogenic of P. aeruginosa7. Upon phagocytosis in cultured macrophages, P. aeruginosa first resides in membrane bound vacuoles, whereas a cytosolic location can be observed at later time of infection7,8. Moreover, ruptured phagosomal membrane has been visualized by TEM in macrophages infected with wild-type P. aeruginosa and a quantitative phagosomal rupture assay has revealed that oprF mutant is less efficient to escape from phagosome than wild-type PAO1 strain8. Here, we showed that oprF mutant in the PAO1 genetic background exhibited a higher association with acidic compartments of cultured macrophages, identified as phagolysosomes by TEM, than the wild-type strain. In addition, a lower number of intracellular bacteria per macrophage was visualized 3 h after infection with the mutant strain compared to wild-type, thus supporting a correlative link between preferential colocalization with acidified compartments and intramacrophage survival defect for oprF mutant. Importantly, the oprF mutant recovered virulence in zebrafish embryo upon treatment with bafilomycin, known to hinder acidification, thus supporting the hypothesis that OprF acts by facilitating bacterial avoidance to macrophage acidification.
The outer membrane protein OprF is involved is cell envelope integrity and abundant amounts of OprF are present in P. aeruginosa outer membrane vesicles (OMVs), from both planktonic cell supernatants and biofilms13,27. In the present study, TEM allowed us to visualize vesicles that are likely of bacterial origin found associated with the phagosomal membrane. The intracellular function, if any, of these vesicles is unknown. Vesicles were also clearly produced from oprF mutant as well, which is consistent with the fact that while OprF is abundant in OMVs, an oprF mutant was shown to produce more OMVs than wild-type in liquid culture35. Thus, we concluded that such vesicles should not be linked to the phenotype of oprF mutant intracellularly, even though we cannot exclude functional differences between OMVs from wild-type strain and oprF mutant.
OprF plays also an important role in the regulation of P. aeruginosa virulence factors. Notably, P. aeruginosa oprF mutant has been shown to reduce expression of T3SS genes and secretion of ExoT and ExoS toxins6,11, which are bi-functional cytotoxins that contain N-terminal RhoGAP domains and C-terminal ADP-ribosylation domains36. Interestingly, similarly to the oprF mutant, a T3SS mutant was attenuated in zebrafish embryos and macrophage depletion restored the virulence of the attenuated T3SS strain21. The in vivo phenotype of oprF mutant could therefore be related to a reduced expression of T3SS genes. Moreover, the phagosomal escape defect of oprF mutant has been attributed to the negative regulation of T3SS expression inside macrophages, since T3SS and more specifically ExoS were found to play a role P. aeruginosa phagosomal escape8. Hence, the stronger association of oprF mutant with acidified compartments may reflect bacterial location in phagolysosome due to a lower ability to escape from the phagosome into the cytosol, in relation with the decreased expression of T3SS genes and reduced secretion of ExoS toxin. Alternatively, OprF, and the T3SS effector ExoS, could play a role in limiting phagolysosomal fusion or acidification of the phagosome. Remarkably and consistent with our findings with macrophages, P. aeruginosa has been proposed to utilize ExoS to avoid acidified compartments within epithelial cells37. Whether ExoS inhibits directly vacuolar acidification in epithelial cells directly, or by redirecting bacteria to other compartments within the cell was not determined.
In summary, based on a combination of animal and cellular experimental approaches, our results indicate that OprF protects P. aeruginosa against macrophage clearance during acute infection, by avoiding destruction in phagolysosomes. The mechanism involved in the avoidance of acidic compartments is not characterized and could involve a facilitation of phagosome escape and/or an impairment of phagosome maturation. Based on previous work, this effect is likely correlated with the intracellular effect of OprF on expression of ExoS, a T3SS effector which has been implicated in phagosomal escape8 and avoidance of acidified compartments in epithelial cells37. Our results also highlight the contribution of macrophages in the clearance of P. aeruginosa during acute infection in the zebrafish embryo model, which appears as the state-of-the-art model to address in vivo the role of macrophages during P. aeruginosa infection.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains and plasmids are described in Table 1. P. aeruginosa was grown at 37 °C in Luria broth (LB). Plasmid pMF230 expressing GFP constitutively38 (obtained from Addgene) was introduced in P. aeruginosa by conjugation, using an E. coli strain containing pRK2013. Recombinant bacteria were selected on LB agar plates containing carbenicillin (300 μg/ml) and triclosan (15 µg/ml).
Infection of Danio rerio embryos
Experiments were performed using the AB zebrafish or the tg(mfap4:mCherry-F) zebrafish line harboring red-fluorescent macrophages25 and maintained under standard conditions6. Bacterial strains, which were freshly streaked out from glycerol stocks, were grown in LB medium to mid-log phase (DO = 0.7 to 0.8), recovered by centrifugation and washed twice in Phosphate-Buffered Saline (PBS). Suspensions were homogenized through a 26-gauge needle and resuspended in PBS at about 109 bacteria/ml added with 10% phenol red to aid visualization of the injection process. Infection were carried by the direct microinjection of 2 nl of bacterial suspensions into the caudal vein of 50 hpf embryos, previously dechorionated and anesthetized with 0.02% tricaine. For survival kinetics after infection, the number of dead embryos was determined visually based on the absence of heartbeat. For phagocytosis visualization, 1500–2000 CFU were injected locally into the hindbrain vesicle (HBV) or the muscle of 50 hpf tg(mfap4:mCherry-F) larvae. To quantify bacteria by CFU counting, infected larvae were dissolved by pipetting in 200 μL 1% PBS 1X-Triton solution at 20 hpi, and plated on LB plates containing ampicillin (100 μg/ml). Depletion of macrophages was carried out upon microinjection of LipoCld or lipoPBS as control into the caudal vein of 24–30 hpf embryos zebrafish24 and visualized by fluorescence microscopy. To inhibit acidification by host vacuolar ATPase, larvae were treated with Bafilomycin A1 (Interchim) at 50 nM in 0.5% DMSO via soaking 30 min before infection in the HBV, as described above29.
Microscopic analysis of zebrafish embryos, quantification of bacterial load by fluorescent pixel counts and quantification of recruited/infected macrophages
For live imaging, anesthetized infected embryos were mounted in 35 mm glass-bottom dishes (FluoroDish, World Precision Instruments, UK) and immobilized with 1% low-melting point agarose. Direct visualization is performed as before6 using an Olympus MVX10 epifluorescent stereomicroscope equipped with a digital color camera (Olympus XC50). Fluorescence and bright-field images are acquired and processed with CellSens (Olympus) and assembled using GIMP 2.6 freeware and Image J software to adjust contrast and brightness and to remove out-of-focus background fluorescence.
For phagocytosis observation, immobilized embryos were immersed with fish water containing tricaine for direct visualization using spinning disc Nikon Ti Andor CSU-W1 microscope (40x/1.15 Water objective).
For time lapse video microscopy of Pseudomonas /macrophage interaction, we used as before39 an ANDOR CSU-W1 confocal spinning disk on an inverted NIKON microscope (Ti Eclipse) with ANDOR Neo sCMOS camera (40 × NA 1.15 water objective). Image stacks for time-lapse movies were acquired at 28 °C every hour, typically spanning 120 μm at 1–2 μm intervals. The 4D files generated from time-lapse acquisitions were processed using Image J, compressed into maximum intensity projections. For quantification of bacterial load by Fluorescent Pixel Counts (FPC), fluorescent bacteria were injected in the larvae and imaged using MVX10 Olympus stereomicroscope or using maximum projection from confocal stacks. Fluorescence was quantified as before39 by computation using Fiji (ImageJ software) as following: 1/ Background was measured in images of PBS injected larvae and then was subtracted in the fluorescence images, 2/ “make binary” function was run, and 3/ “measure area” function was used to determine the number of fluorescent pixels of the image, with avoiding the auto-fluorescence of the yolk.
Recruited and infected macrophages were counted manually from z-stacks of obtained microscopy images, using Fiji (ImageJ) software, after the brightness/contrast adjustment for better visualization. Macrophages were considered as infected if macrophage fluorescence overlaps with bacterial fluorescence in respective z-stack.
Infection of cultured macrophages and visualization and quantification of intracellular bacteria by fluorescent microscopy
J774A.1 cells were maintained at 37 °C in 5% CO2 in Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco). The infection of J774 macrophages seeded on glass coverslips by P. aeruginosa was carried out as described previously6. After 2.5 h of gentamicin treatment, the cells were washed twice with PBS and fixed with 4% paraformaldehyde for 30 min. To visualize acidified compartments, macrophages were incubated with 50 nM Lysotracker red DND-99 (Molecular Probes) in DMEM (supplemented with 10% FBS) for 10–15 min before fixation to stain lysosomes exclusively. After fixation, cells were washed, mounted on glass slides in Vectashield with DAPI (Vector Laboratories, Inc) and slides were examined as described previously8 using an upright fluorescence microscope (Axioimager Z2, Zeiss) equipped with an Apotome 1 for optical sectioning. A 63X Apochromat Objective (NA 1.4). Transmitted light was acquired using differential interference contrast (DIC), Fluorescein isothiocyanate (FITC) filter was used to visualize GFP expressing bacteria and Lysotracker red fluorescence was acquired using a texas red filter set. Images were processed using ZEN blue software (Zeiss).
Transmission electron microscopy
Macrophages were seeded on glass coverslips and infected as described above. Infected cells were fixed with 2.5% gluteraldehyde and treated as described previously8. Dehydration was performed through acetonitrile series and samples impregnated in epon 118: acetonitrile 50:50, followed by two times for 1 h in 100% epon, were treated as described8. Ultrathin sections of 70 nm were cut with a Leica UC7 ultramicrotome (Leica microsystems), counterstained with uranyl acetate and lead citrate and observed in a Jeol 1200 EXII transmission electron microscope. All chemicals were from Electron Microscopy Sciences (USA) and solvents were from Sigma. Images were processed using Fiji software.
Ethics statement
All animal experiments described in the present study were conducted at the University of Montpellier according to European Union guidelines for handling of laboratory animals (http://ec.europa.eu/environment/chemicals/lab_animals/home_en.htm) and were approved by the Direction Sanitaire et Vétérinaire de l'Hérault and Comité d'Ethique pour l'Expérimentation Animale under reference CEEA-LR-B4-172-37 and APAFIS#5737-2016061511212601 v3. The breeding of adult fish adhered to the international guidelines specified by the EU Animal Protection Directive 2010/63/EU and adult zebrafish were not sacrificed for this study. All experiments were performed before the embryos free-feeding stage and did not fall under animal experimentation law according to the EU Animal Protection Directive 2010/63/EU. For survival curves, cardiac rhythm was used as a clinical criterium. Embryos were euthanized using the anaesthetic Tricaine up to a lethal dose (500 mg/ml) before bleach treatment.
Statistical analysis
Statistical analyses for ex vivo experiments with J774 cells was performed using t-test and comparisons between survival curves were performed using the log rank test with Prism 5.01 (GraphPad, Inc.). Statistical analysis for macrophage recruitment and phagocytosis is indicated in the legend of Fig. 2. In the figures, * means p value ≤ 0.05, ** ≤ 0.01, *** ≤ 0.001 and **** ≤ 0.0001.
References
Mittal, R. et al. In vitro interaction of Pseudomonas aeruginosa with human middle ear epithelial cells. PLoS ONE 9, e91885. https://doi.org/10.1371/journal.pone.0091885 (2014).
Kazmierczak, B. I., Jou, T. S., Mostov, K. & Engel, J. N. Rho GTPase activity modulates Pseudomonas aeruginosa internalization by epithelial cells. Cell Microbiol. 3, 85–98 (2001).
Angus, A. A. et al. Pseudomonas aeruginosa induces membrane blebs in epithelial cells, which are utilized as a niche for intracellular replication and motility. Infect. Immun. 76, 1992–2001. https://doi.org/10.1128/IAI.01221-07 (2008).
Kroken, A. R., Chen, C. K., Evans, D. J., Yahr, T. L. & Fleiszig, S. M. J. The impact of ExoS on Pseudomonas aeruginosa internalization by epithelial cells is independent of fleQ and correlates with bistability of type three secretion system gene expression. mBio https://doi.org/10.1128/mBio.00668-18 (2018).
Buyck, J. M., Tulkens, P. M. & Van Bambeke, F. Pharmacodynamic evaluation of the intracellular activity of antibiotics towards Pseudomonas aeruginosa PAO1 in a model of THP-1 human monocytes. Antimicrob. Agents Chemother. 57, 2310–2318. https://doi.org/10.1128/AAC.02609-12 (2013).
Belon, C. et al. A macrophage subversion factor is shared by intracellular and extracellular pathogens. PLoS Pathog. 11, e1004969. https://doi.org/10.1371/journal.ppat.1004969 (2015).
Mittal, R. et al. Otopathogenic Pseudomonas aeruginosa enters and survives inside macrophages. Front. Microbiol. 7, 1828. https://doi.org/10.3389/fmicb.2016.01828 (2016).
Garai, P., Berry, L., Moussouni, M., Bleves, S. & Blanc-Potard, A. B. Killing from the inside: Intracellular role of T3SS in the fate of Pseudomonas aeruginosa within macrophages revealed by mgtC and oprF mutants. PLoS Pathog. 15, e1007812. https://doi.org/10.1371/journal.ppat.1007812 (2019).
Bernut, A., Belon, C., Soscia, C., Bleves, S. & Blanc-Potard, A. B. Intracellular phase for an extracellular bacterial pathogen: MgtC shows the way. Microb. Cell 2, 353–355. https://doi.org/10.15698/mic2015.09.227 (2015).
Fito-Boncompte, L. et al. Full virulence of Pseudomonas aeruginosa requires OprF. Infect. Immun. 79, 1176–1186. https://doi.org/10.1128/IAI.00850-10 (2011).
Bouffartigues, E. et al. The absence of the Pseudomonas aeruginosa OprF protein leads to increased biofilm formation through variation in c-di-GMP level. Front. Microbiol. 6, 630. https://doi.org/10.3389/fmicb.2015.00630 (2015).
Chevalier, S. et al. Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol. Rev. 41, 698–722. https://doi.org/10.1093/femsre/fux020 (2017).
Cassin, E. K. & Tseng, B. S. Pushing beyond the envelope: the potential roles of OprF in Pseudomonas aeruginosa biofilm formation and pathogenicity. J. Bacteriol. https://doi.org/10.1128/JB.00050-19 (2019).
Kooguchi, K. et al. Role of alveolar macrophages in initiation and regulation of inflammation in Pseudomonas aeruginosa pneumonia. Infect. Immun. 66, 3164–3169 (1998).
Cheung, D. O., Halsey, K. & Speert, D. P. Role of pulmonary alveolar macrophages in defense of the lung against Pseudomonas aeruginosa. Infect. Immun. 68, 4585–4592. https://doi.org/10.1128/iai.68.8.4585-4592.2000 (2000).
Masud, S., Torraca, V. & Meijer, A. H. Modeling infectious diseases in the context of a developing immune system. Curr. Top. Dev. Biol. 124, 277–329. https://doi.org/10.1016/bs.ctdb.2016.10.006 (2017).
Torraca, V. & Mostowy, S. Zebrafish infection: from pathogenesis to cell biology. Trends Cell Biol. 28, 143–156. https://doi.org/10.1016/j.tcb.2017.10.002 (2018).
Rosowski, E. E. Illuminating macrophage contributions to host-pathogen interactions in vivo: the power of Zebrafish. Infect. Immun. https://doi.org/10.1128/IAI.00906-19 (2020).
Yoshida, N., Frickel, E. M. & Mostowy, S. Macrophage–microbe interactions: lessons from the zebrafish model. Front. Immunol. 8, 1703. https://doi.org/10.3389/fimmu.2017.01703 (2017).
Torraca, V., Masud, S., Spaink, H. P. & Meijer, A. H. Macrophage-pathogen interactions in infectious diseases: new therapeutic insights from the zebrafish host model. Dis. Model. Mech. 7, 785–797. https://doi.org/10.1242/dmm.015594 (2014).
Brannon, M. K. et al. Pseudomonas aeruginosa Type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell Microbiol. 11, 755–768. https://doi.org/10.1111/j.1462-5822.2009.01288.x (2009).
Kumar, S. S. et al. Dual transcriptomics of host–pathogen interaction of cystic fibrosis isolate Pseudomonas aeruginosa PASS1 With Zebrafish. Front. Cell. Infect. Microbiol. 8, 406. https://doi.org/10.3389/fcimb.2018.00406 (2018).
van Rooijen, N., Sanders, A. & van den Berg, T. K. Apoptosis of macrophages induced by liposome-mediated intracellular delivery of clodronate and propamidine. J. Immunol. Methods 193, 93–99. https://doi.org/10.1016/0022-1759(96)00056-7 (1996).
Bernut, A. et al. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc. Natl. Acad. Sci. USA 111, E943-952. https://doi.org/10.1073/pnas.1321390111 (2014).
Phan, Q. T. et al. Neutrophils use superoxide to control bacterial infection at a distance. PLoS Pathog. 14, e1007157. https://doi.org/10.1371/journal.ppat.1007157 (2018).
Colucci-Guyon, E., Tinevez, J. Y., Renshaw, S. A. & Herbomel, P. Strategies of professional phagocytes in vivo: unlike macrophages, neutrophils engulf only surface-associated microbes. J. Cell Sci. 124, 3053–3059. https://doi.org/10.1242/jcs.082792 (2011).
Couto, N., Schooling, S. R., Dutcher, J. R. & Barber, J. Proteome profiles of outer membrane vesicles and extracellular matrix of Pseudomonas aeruginosa biofilms. J. Proteome Res. 14, 4207–4222. https://doi.org/10.1021/acs.jproteome.5b00312 (2015).
Bowman, E. J., Siebers, A. & Altendorf, K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA 85, 7972–7976. https://doi.org/10.1073/pnas.85.21.7972 (1988).
Levitte, S. et al. Mycobacterial acid tolerance enables phagolysosomal survival and establishment of tuberculous infection in vivo. Cell Host Microbe 20, 250–258. https://doi.org/10.1016/j.chom.2016.07.007 (2016).
Gomes, M. C. & Mostowy, S. The case for modeling human infection in Zebrafish. Trends Microbiol. 28, 10–18. https://doi.org/10.1016/j.tim.2019.08.005 (2020).
Hernandez, Y. L., Yero, D., Pinos-Rodriguez, J. M. & Gibert, I. Animals devoid of pulmonary system as infection models in the study of lung bacterial pathogens. Front. Microbiol. https://doi.org/10.3389/Fmicb.2015.00038 (2015).
Clatworthy, A. E. et al. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect. Immun. 77, 1293–1303. https://doi.org/10.1128/IAI.01181-08 (2009).
Cafora, M. et al. Phage therapy against Pseudomonas aeruginosa infections in a cystic fibrosis zebrafish model. Sci. Rep. 9, 1527. https://doi.org/10.1038/s41598-018-37636-x (2019).
Mishra, M., Ressler, A., Schlesinger, L. S. & Wozniak, D. J. Identification of OprF as a complement component C3 binding acceptor molecule on the surface of Pseudomonas aeruginosa. Infect. Immun. 83, 3006–3014. https://doi.org/10.1128/IAI.00081-15 (2015).
Wessel, A. K., Liew, J., Kwon, T., Marcotte, E. M. & Whiteley, M. Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. J. Bacteriol. 195, 213–219. https://doi.org/10.1128/JB.01253-12 (2013).
Barbieri, J. T. & Sun, J. Pseudomonas aeruginosa ExoS and ExoT. Rev. Physiol. Biochem. Pharmacol. 152, 79–92. https://doi.org/10.1007/s10254-004-0031-7 (2004).
Heimer, S. R. et al. Pseudomonas aeruginosa utilizes the type III secreted toxin ExoS to avoid acidified compartments within epithelial cells. PLoS ONE 8, e73111. https://doi.org/10.1371/journal.pone.0073111 (2013).
Nivens, D. E., Ohman, D. E., Williams, J. & Franklin, M. J. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183, 1047–1057. https://doi.org/10.1128/JB.183.3.1047-1057.2001 (2001).
Phan, Q. T. et al. Neutrophils use superoxide to control bacterial infection at a distance. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1007157 (2018).
Acknowledgements
We thank Sylvie Chevalier (Rouen University) for providing strains and Audrey Bernut (Versailles Saint-Quentin University) for critical reading of the manuscript. We thank the Montpellier RIO Imaging microscopy platform, member of the national infrastructure France-BioImaging infrastructure supported by the French National Research Agency (ANR-10-INBS-04, «Investments for the future») for photonic Microscopy and the Electron Microscopy facility of the University of Montpellier (MEA) for sample preparation and transmission electron microscopy. This work is supported by Vaincre La Mucoviscidose (RIF20170502042) and Association Gregory Lemarchal. MM is supported by Vaincre La Mucoviscidose (RIF20170502042). TS is supported by grant from the European Community's H2020 Program [Marie-Curie Innovative Training Network ImageInLife: Grant Agreement No. 721537].
Author information
Authors and Affiliations
Contributions
M.M., M.N-C. and A-B.B-P. conceived and designed experiments. M.M. and T.S. performed experiments. L.B. performed and analyzed TEM. All authors contributed in formal analysis. M.M. and A-B.B-P. wrote the manuscript. All authors edited the manuscript and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Moussouni, M., Berry, L., Sipka, T. et al. Pseudomonas aeruginosa OprF plays a role in resistance to macrophage clearance during acute infection. Sci Rep 11, 359 (2021). https://doi.org/10.1038/s41598-020-79678-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-79678-0
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.