Abstract
The Human Developmental Cell Atlas (HDCA) initiative, which is part of the Human Cell Atlas, aims to create a comprehensive reference map of cells during development. This will be critical to understanding normal organogenesis, the effect of mutations, environmental factors and infectious agents on human development, congenital and childhood disorders, and the cellular basis of ageing, cancer and regenerative medicine. Here we outline the HDCA initiative and the challenges of mapping and modelling human development using state-of-the-art technologies to create a reference atlas across gestation. Similar to the Human Genome Project, the HDCA will integrate the output from a growing community of scientists who are mapping human development into a unified atlas. We describe the early milestones that have been achieved and the use of human stem-cell-derived cultures, organoids and animal models to inform the HDCA, especially for prenatal tissues that are hard to acquire. Finally, we provide a roadmap towards a complete atlas of human development.
Similar content being viewed by others
Main
Most modern developmental biology research has historically focused on model organisms. Owing to practical challenges, human development—from a fertilized ovum to a fully formed fetus at birth—has remained a poorly understood ‘black box’. The implications of a human developmental cell atlas for understanding human development are far-reaching, as many congenital disorders and childhood cancers may originate during susceptible windows of development1,2,3. The clinical relevance of the atlas extends into adulthood for ageing, cancer and applications in regenerative medicine and stem cell therapies4,5,6. Furthermore, embryonic and fetal stem cells7,8 and developmental trajectories provide an essential reference and guide for engineering human stem-cell-derived models9,10,11,12,13, organoids14 and cellular therapies.
Human development begins with a fertilized oocyte that divides and differentiates through preimplantation, embryonic and fetal stages (Fig. 1). Early studies began with morphometric and qualitative assessments of human embryos, leading to development of the Carnegie staging system15 (Fig. 1). Advances in imaging, cytometry and genomics technologies have provided further insights into the complex spatiotemporal changes during organogenesis16. Recent progress in single-cell profiling technologies has revolutionized our ability to study human development at an unprecedented resolution17. Leveraging these advances to build a comprehensive atlas of human development (from the fertilized oocyte to birth) at cellular resolution is an ambitious endeavour that is similar in scale to the Human Genome Project, which required multidisciplinary scientific expertise from disparate fields working together collaboratively. Such a community has arisen from the grassroots assembly of global researchers who are working as part of the Human Cell Atlas (HCA)18 initiative. As with the Human Genome Project, the HCA will be a foundational scientific resource, composed of diverse data types and available freely through browsable and searchable web portals that visualize cells across anatomical space and developmental time.
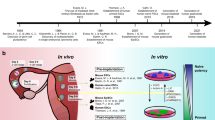
a, Timeline of human development from fertilization to birth. CT, cytotrophoblast; ESC, embryonic stem cell; EPI, epiblast; PrE, primitive endoderm; pcw, post-conception weeks; SCT, syncytiotrophoblast; TE, trophectoderm; TSC, trophoblast stem cell; XEN, extraembryonic endoderm; YSE, yolk sac endoderm. b, Models derived from human stem cells, and associated studies. In vitro model systems to study early embryonic development. A–P, anterior–posterior. c, Experimental model systems to study development, including Drosophila melanogaster, Danio rerio, Xenopus laevis, Gallus gallus, Mus musculus, cell culture and organoids, and their amenability to facilitating various aspects of scientific study.
The HDCA is a strategic focus of HCA19, and is pursued by scientists from individual laboratories as well as large national and international research consortia (Supplementary Table 1); the HDCA is open to all who adhere to its mission and open science values20. The HDCA aims for equity, inclusivity and diversity both in terms of scientific participation and the representation of human tissue samples. We encourage any interested researcher to become a member, participate, register their study and contribute their data and publications to the HDCA and HCA21.
Building a developmental cell atlas
The successful construction of a HDCA poses substantial scientific challenges in terms of experimental measurement technologies, computational analysis and visualization algorithms (Fig. 2). In particular, the dynamic nature of gestation creates challenges for designing a sampling strategy, especially to capture transient morphological changes in the first eight weeks. A major endeavour for the HDCA will be to develop the conceptual and computational framework to capture development with respect to cellular and morphological changes. The HDCA, through coordination with the HCA Organoid Network22, will incorporate data from in vitro culture model and organoid systems23 to cautiously infer development between seven days and four weeks after conception (a period in which samples are difficult to obtain) (Fig. 1b, c).
a, ‘How to build an atlas’ modules, including an interdisciplinary team (biological disciplines, clinical specialities, computational scientists and technology engineers), multimodal technologies and integration of data across platforms. b, Key features of the HDCA. Single-cell measurements across three-dimensional space (alongside a fourth dimension of time) allow for the capture of dynamic developmental processes, including cell proliferation, migration and regulation. Spatial extension captures cell proliferation (changes in organ size and shape), and the sensing and control of morphogenesis; spatio-temporal events include cell differentiation and replication, and changes in cellular composition and gene expression. Permutations and variabilities outline the spatial determinants of cell differentiation, mechanical adhesive feedback and heterocellular signalling by ligands. Time series trajectories reveal fate-biased migrating progenitors, lineage specifications and cell migration. c, Utility and applications of the HDCA: cellular and molecular biological insights into the coordination of organ development across the whole embryo (left) are applied to advance regenerative medicine, tissue engineering and therapeutic strategies (right).
The successful delivery of the HDCA will leverage the Human Genome Project-initiated restructuring of how large science projects are funded, conducted, coordinated and shared (based on the Fort Lauderdale Principles24) that forms the basis for the HCA, its committees (for example, computation and ethics) and ‘Biological Networks’20. This organizational framework has enabled researchers to form large-scale coordinated collaborations across technologies and biological disciplines: developmental biology, embryology, genetics and model systems, computational biology, clinical specialities (including in vitro fertilization), clinical genetics and pathology, as well as coordination with funders. Partnerships with allied biological networks, including organoid and paediatric atlas projects, will facilitate clinical applications.
Ethics, resources and data sharing
Accessing human developmental samples is constrained by general and geographically specific ethical and legal challenges. These include issues relating to donation, access to and research use of legally defined developing human tissue material, regulatory approvals processes and cultural sensitivities. Research on human embryos and fetuses is supported within European and national regulations, such as the UK National Research Ethics Service (NRES) and the French Agence de Biomédecine. In the UK, studies on preimplantation human embryos (up to 14 days after conception) are governed by the Human Fertilisation and Embryology Authority and a research ethics committee (such as NRES). However, in the USA, research on donated human embryonic and fetal materials has increasingly been restricted over the past two decades, despite the existence of similar regulatory oversight.
Nonetheless, resources to support research in human development (such as the UK’s Human Developmental Biology Resource (HDBR)25) provide material to researchers. Recipients of HBDR tissue who are not based in the UK require their own project-specific ethics approval, before receipt of material. The HDBR provides embryonic and fetal samples from 4 to 20 weeks after conception with karyotype information and, increasingly, with anonymized maternal DNA and clinical history. Material from fetuses with prenatally diagnosed disorders is also available. The French Human Developmental Cell Atlas (HuDeCA) (https://hudeca.genouest.org) has recently been established, and aspires to constitute a comprehensive European resource of human embryonic or early fetal samples.
The international sharing of genomic sequencing and clinical data derived from prenatal or paediatric tissue samples is subject to data protection regulation that considers live versus deceased status, consent regarding research data use and confidentiality. Data from living donors are shared under appropriate access controls. The HCA Ethics Working Group is developing tools, guidance notes26, consent-form templates and sampling information for embryonic, fetal and paediatric tissue material, and international data-sharing guidance for the HDCA.
Mapping development across space and time
Development is intricately orchestrated in three spatial dimensions and gestation time. Human embryogenesis cannot be easily assessed at high resolution in vivo27. Time-lapse studies are limited to in vitro preimplantation embryos. The application of high-throughput genomics technologies to dissociated cells and tissue sections in situ is beginning to provide data of unprecedented resolution (Fig. 3, Table 1).
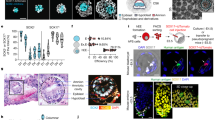
a, Organ or anatomical unit profiling of a prenatal embryo derived from multiple germ layers. b, Single-cell atlas technologies by relative resolution and genome scale. CODEX, co-detection by indexing; H&E, haematoxylin and eosin staining; IF, immunofluorescence; ISS, in situ sequencing; MERFISH, multiplexed error-robust FISH; MIBI, multiplexed ion beam imaging; seqFISH, sequential FISH; smFISH, single-molecule FISH; STARmap, spatially resolved transcript annotation readout mapping. c, Integration of datasets from different technologies (for example, spatial transcriptomics, scRNA-seq and targeted ISS) to profile organs or whole embryos.
Cellular and molecular heterogeneity
Single-cell molecular profiles based on RNA, chromatin accessibility, methylation or select protein signatures have enabled a more nuanced definition of cell types and states. The data underpinning such definitions are increasingly derived from single-cell RNA sequencing (scRNA-seq), barcoded antibodies and accessible chromatin sequencing of dissociated cells28,29. Resolving cell types and trajectories at high granularity is aided by full-length scRNA-seq, but is primarily performed by profiling large numbers of cells. Cell-type definition is currently guided by existing knowledge from model organisms and adult cellular profiles, which may not faithfully reflect prenatal cell types, transient cell types that are present only during development and transitional states of differentiation.
To overcome these challenges, many time points need to be profiled and defined cell states need to be mapped back into their 3D space over time and functionally characterized. High levels of multiplexing can attain this level of granularity at an affordable cost for a complete human developmental cell atlas30,31. Molecular profiles, morphology, functional assessment and other features can reflect the multifaceted state of a cell. For example, the transcriptome reflects the present and potential future of a cell; protein expression captures the immediate past and present state of a cell; chromatin profiles reveal its invariant type and potential for future differentiation; and ontogeny reveals its history.
The field of developmental biology has traditionally drawn on ontogenic relationships to define cell types, but this is challenging in humans for whom information is captured as snapshots across gestation. CRISPR scarring is applicable only in stem cells, organoid systems and short-term explants32,33. The tracking of somatic mutations is the only available technology to definitively determine ontogeny, but is limited by its current lack of scalability34,35. Recent methods that rely on the simultaneous measurement of mitochondrial DNA and RNA, transcriptome and open chromatin may overcome this challenge36,37. We anticipate the field moving towards a consensus cell ontology that integrates multimodal single-cell profiling data as well as legacy knowledge of embryonic cell-type definitions augmented by information from diverse animal models.
Mapping cells in 2D and 3D
Spatial genomics methods to measure RNA in tissue sections typically offer a trade-off: high-resolution (single-cell and subcellular) methods that typically measure hundreds of transcripts or whole transcriptome profiles at a multicellular level38,39. This trade-off can be mitigated by integration with single-cell profiles from dissociated cells, expanding the genomic coverage by predicting the spatial expression of unmeasured genes or enhancing resolution by deconvolution of multicellular measurements. Tissue clearing methods to render organs transparent40, combined with whole-mount protein immunostaining and RNA single-molecule fluorescence in situ hybridization (FISH)41,42, can now provide 3D molecular profiling at cellular or subcellular resolution using light-sheet microscopy43,44,45. Increasing multiplex capacity and use of artificial intelligence and machine learning algorithms to overcome data analytical challenges have successfully been deployed to image whole-organismal vasculature following tissue clearing46,47.
Biophysical methods and live imaging
Mounting evidence from Drosophila and other models shows that mechanical forces have a key role in development processes and tissue morphogenesis48. Surface tension and pressure can be measured in single cells of preimplantation mouse embryos49. Adapting these technologies to human preimplantation embryos and stem-cell-based embryo models50 can build a spatiotemporal mechanical atlas.
Positional landmarks in development
A standard coordinate system for locations in the human body (a common coordinate framework (CCF)) is crucial for the HCA and HDCA51. Two types of systems are useful: absolute (similar to postcode or zip code addresses) and relative (similar to a landmark-based address system). CCF anatomical ‘postcodes’ enable the integration of multimodal datasets of different spatial and longitudinal resolution. The Allen Mouse Brain Reference Atlas version 3 provides a CCF of 3D anatomical features and local features grouped in a hierarchy to facilitate multilevel analysis of the mouse brain. Efforts are currently underway to establish CCFs for adult human organs within the Human Biomolecular Atlas Program of the National Institutions of Health (NIH). The HDCA will need to develop a CCF that incorporates space and time, as well as cell movement and patterns during organogenesis on the basis of existing macro-level 3D coordinates for human embryos (such as the HDBR atlas (http://hdbratlas.org/) and the Transparent Human Embryo (https://transparent-human-embryo.com/)).
Computation and data visualization
Among the key algorithmic challenges to integrating data into a developmental atlas are (1) mapping cells with more intermediate states compared to adult counterparts; (2) inferring time orderings and lineage relations, including branching lineages and multiple paths converging on the same outcome; (3) inferring spatial movement of cells; (4) building a temporal series of CCFs, each as a probabilistic model for a time window as well as a model for their morphing along space and time52; (5) mapping across modalities and time points (for example, chromatin states in one time window to RNA and protein levels of another); and (6) regulatory and molecular network inference within and across cells. Theories and insights from multiple fields will be required to model the mechanisms that underpin tissue formation and growth. It is likely that additional emergent properties of cells and their ecosystems will be discovered using interdisciplinary approaches. These will need new vocabularies, ontologies and modelling approaches to be understood. The HDCA community must also apply FAIR (findability, accessibility, interoperability and reusability) principles to help to ensure reproducibility and data accessibility53.
Computational integration of multi-omics data for visualizations similar to that of Google Maps, such as the Open Microscopy Environment (https://www.openmicroscopy.org/), will enable zooming to the single-cell level from a large-volume tissue view. Additional complexity will combine visualizations from imaging and sequencing data. A sophisticated abstraction of raw data and integration across modalities, anchored by a developmental CCF, will be essential. Links to clinical relevance and applications will enhance the utility of the atlas.
Emerging cell atlases of human development
The advantages of whole tissue or organ profiling compared to lineage-centric analysis include comprehensive cellular analysis and the discovery of emergent biological properties. For example, the developing liver functions as a haematopoietic organ during early gestation until the middle of the second trimester, before it functionally transitions into a metabolic organ similar to the adult liver54. To meet the high demand for erythropoiesis during development, the human skin and adrenal glands can also support erythrocyte maturation during the first trimester54,55.
In contrast to our terrestrial postnatal life, the human embryo and fetus exist in an aquatic environment: our lung, gut and skin are exposed to amniotic fluid. In contrast to the postnatal lung, the developing lung does not perform oxygen transfer or receive the same volume of blood through the pulmonary veins. The effect of these physiological factors on individual tissues and the role of the placenta and maternal decidua in supporting human embryogenesis and fetal life are emerging56,57.
Current organ atlases of brain, gut, heart, liver, kidney, placenta, thymus and skin (Table 1) underscore the importance of studying human samples and have revealed unique aspects of human development that are not conserved with animal model systems58,59,60,61. These include timelines of development during gestation, cell-type markers and the expression pattern of transcription factors between mouse and human organs62,63.
The specification of functional tissue niches occurs during both prenatal and postnatal life. Studies of the fetal gut have highlighted the importance of interactions between the epithelial and mesenchymal compartments in allowing the formation of villi, and have identified fetal gut transcription factors that are aberrantly activated in individuals with paediatric Crohn’s disease64. Comparison between the developing and adult kidney have demonstrated the establishment of a dedicated spatial zonation pattern that protects against uropathogenic bacterial challenges postnatally61,65. Single-cell transcriptomics of germ cells during development have provided important insights into the main pathways that control their differentiation66,67, with ongoing studies focused on exploring the regulatory mechanisms of sex determination (https://hugodeca-project.eu).
Early developmental studies of the brain have focused on human and primate cortical development68,69,70. The developing human and rodent midbrain, which contains the clinically relevant dopaminergic cell groups that are lost in individuals with Parkinson’s disease, has also extensively been studied63,71,72, as have the developing mouse spinal cord and cerebellum73,74, the hypothalamic arcuate nucleus and the diencephalon75.
Atlases of distributed systems (such as the immune system) have been initiated, detailing haematopoietic organs such as the yolk sac76,77 and liver54, lymphoid tissues such as thymus (in which T cells differentiate)78 and non-lymphoid tissues such as skin and kidney, in which immune cells reside. These studies have revealed an intrinsic change in the differentiation potential of haematopoietic stem progenitor cells with gestational time, together with the importance of the local tissue microenvironment for blood and immune-cell development.
Model organisms and culture systems
Our understanding of human development has largely been inferred from studies on animal model systems that are not always conserved across species79 (Fig. 1). Two recent studies contrast the kinetics of development between human and mouse, highlighting the need for caution in interpreting heterospecific graft studies and findings from nonprimate preclinical models80,81. However, the feasibility of perturbation and in-depth mechanistic studies using animal models and culture systems provide a valuable scaffold and complement the HDCA, particularly for the immediate weeks after implantation during which human samples are inaccessible.
Single-cell molecular profiling has transformed many aspects of developmental biology research across all major model organisms82,83,84,85,86, providing mechanistic insights into fundamental biological processes (including the early specification of germ layers and diversification of early cardiovascular cells)29,87. Comparative biology has the potential to make major contributions to cell ontology. The availability of parallel human and model species data will support expanded cross-species analyses. Computational analysis can align cells and inferred lineages across species to extrapolate findings from nonprimate models and help to optimize animal models of normal and pathological human development. From a computational perspective, it will be important to develop tools for better annotation of 3′ and 5′ untranslated regions of animal model data, as most scRNA-seq technologies capture only these regions. The development of computational tools that can robustly map developmental trajectories across species and that can account for different developmental kinetics between cell types within and between species will be required. Comparative studies of human and mouse preimplantation and gastrulation embryos have revealed conserved and divergent transcriptional programs. For example, Klf2 expression in mouse embryo-fated epiblast progenitor cells is not observed in humans; by contrast, KLF17 is enriched in human, but not mouse, epiblast88.
The self-organization of human embryonic tissue can be captured from the earliest moments in vitro50,89, and extended to gastrulation, anterior–posterior embryonic patterning and the early phases of somitogenesis11. The recent human gastrulation embryo dataset will be informative as a benchmark to further refine in vitro directed differentiation of human cells, including gastruloid models11. Other processes during organogenesis can also be monitored, including the clock control of somite segmentation90,91, boundary formations during hepato-biliary–pancreatic organ budding92 and patterning of the neural tube. Protocols are now established to mimic the development of diverse human tissues that exhibit morphologies and physiological functionalities of developing human tissues. These organoid systems include hair-bearing skin93; the small intestine with a crypt–villus axis94; region-specific95 and multiregion96 brain tissue that models neurogenesis, neural migration and synapse formation; multilayered neural retina with photoreception responses97; and arterio-venous specification during blood vessel development98.
A comprehensive reference atlas of the cell types and states that are present during human development will be critical to benchmark stem-cell-derived organoids. Such roadmap comparisons will highlight similarities69 and deficiencies99, and define strategies for improving organoids for disease modelling. In the future, high-fidelity human stem-cell-derived human organoids and single-cell multi-omic modalities will be powerful tools to understand the mechanisms that control human organogenesis.
Clinical relevance and applications
The interaction of genotype and environment that leads to phenotype underlies developmental disorders. A range of childhood and adult disorders have their origins in prenatal life (Fig. 4). These include structural birth defects100, neurodevelopmental disorders (including schizophrenia)101, childhood cancers2,65, inborn errors of immunity102, infertility and differences of sex development103, as well as many paediatric disorders104. Thousands of rare genetic diseases can each present a spectrum of perturbed developmental sequelae at birth, and sometimes differ widely in medical presentation even when classified as the same disease or condition105. As examples, Down syndrome (trisomy 21)106 and 22q11.2 deletion syndrome107 separately present substantial risks for schizophrenia, Alzheimer’s disease and hypothyroidism starting in adolescence108. Identifying the aetiology of developmental disorders and the effects of maternal genotype, paternal age and other external risk factors (such as diet, alcohol, toxins, endocrine disruptors and pathogens) has been hampered by our limited understanding of normal development in humans.
a, Diseases related to brain development, across lifespan. A timeline of brain development across human life, with examples of diseases with onset at different gestational stages and ages. b, Disease state compared to developmental atlas, showing how a single-cell atlas with temporal and spatial information can be used as a reference to understand disease states.
Development atlases are also revealing the pathogenesis of childhood cancers (Fig. 4). Paediatric and adult brain tumours in their early stages often present impaired developmental programs within tumour cells109,110. Comparing the expression profile of tumour cells with the HDCA can identify the cancer cell of origin and its oncogenic pathways. For example, a single-cell atlas of the developing mouse cerebellum has been used to investigate subtypes of human medulloblastoma (a paediatric brain tumour)2,111, and cell states during nephrogenesis revealed the developmental cellular origin of Wilms’ tumour65. High-resolution mapping of developing immune cells will inform the molecular basis and extent of disease phenotypes of childhood leukaemias and primary immunodeficiencies.
Many adult cancers also recapitulate a dysregulated version of human developmental programs112. The acquisition of early developmental molecular programs is characteristic of malignant pathology, and is a previously unrecognized hallmark of immunological disease and the cancer immune environment113,114. HDCA data have also facilitated our understanding of the differential susceptibility of adult and prenatal cells to SARS-CoV-2 through examination of viral entry receptor and protease expression in a wide range of organs115.
Cell and tissue engineering for clinical therapies and regenerative medicine are areas with considerable potential for the direct utility of the HDCA. Cell therapies derived from human pluripotent stem cells are now entering early clinical trials for the treatment of Parkinson’s disease116, using protocols that were refined on the basis of developmental studies of midbrain dopaminergic neurons72. Similar approaches are being followed to develop a range of other stem cell products for human trials117. Haematopoietic stem cell transplantation is an established and widely used treatment for many haematological, and increasingly non-haematological, disorders. Leveraging the potency factors of fetal haematopoietic stem cells could have a substantial benefit for patients who receive transplants of haematopoietic stem cells.
Towards a whole embryo atlas
The initial HCA white paper emphasized 12 distinct organ systems within the human body and highlighted the importance of a developmental cell atlas. Integrated multi-organ analyses will provide insights into the tissue microenvironment that shapes resident epithelial, stroma and immune cells and the cellular heterogeneity of innervating blood vessels, lymphatics and peripheral nerves. Eventually, this may illuminate system-level lineage development and cell fate decisions across an entire organism. The datasets from profiling based on human developmental organs have been critical in interpreting recent multi-organ developmental atlases55,118.
There are several large-scale organ-based studies being undertaken by HDCA researchers. These include the NIH ‘Brain Research through Advancing Innovative Neurotechnologies’ (BRAIN) initiative (including the BRAIN Initiative Cell Census Network (BICCN) consortium) focusing on the developing human cortex; the Swedish Human Cell Atlas consortium performing large-scale scRNA-seq, assay for transposase-accessible chromatin using sequencing (ATAC-seq) and spatial-omic analyses of the developing human brain, heart119 and lung during the first trimester; the French HuDeCA consortium mapping eight first-trimester human organs using 3D imaging and scRNA-seq; the European Union (EU) Horizon 2020-funded developing brain (Braintime) and gonad (HUGODECA) projects; the NIH Developmental Genotype-Tissue Expression (dGTEx)120 project; and UK consortia funded by the Wellcome Trust and Medical Research Council. The logical next step will be to coordinate these efforts and extend the current approach to contextualize the development of different cell lineages across all organs.
However, multi-organ approaches do not permit the analysis of distributed tissue networks as a continuum from a single donor sample. Whole-embryo analysis has been limited to very early preimplantation samples88,121,122 and one gastrulation-stage embryo123. Multi-omics suspension and spatial-genomics profiling of anatomically dissected units from whole human embryos at six to seven weeks after conception are being undertaken by the HDCA researchers based in the UK. We anticipate a first whole human embryo profiling within the next two years. On the basis of existing HDCA data and the rapid changes during early development, we propose a minimum of three replicates for each biologically relevant gestation period (for example, each week from six weeks after conception). All such data produced and shared by the global research community (formally registered with the HCA or not) contributes to the HDCA. Defining a universal organizing framework for these data will enable them to be unified into a complete atlas that will be a transformative resource for the research and clinical communities.
References
Behjati, S., Lindsay, S., Teichmann, S. A. & Haniffa, M. Mapping human development at single-cell resolution. Development 145, dev152561 (2018).
Vladoiu, M. C. et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 572, 67–73 (2019).
Velmeshev, D. et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689 (2019).
Gulsuner, S. et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529 (2013).
Simmons, R. A. Developmental origins of adult disease. Pediatr. Clin. North Am. 56, 449–466 (2009).
Laughney, A. M. et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat. Med. 26, 259–269 (2020).
Sozen, B., Jorgensen, V., Zhu, M., Cui, T. & Zernicka-Goetz, M. Reconstructing human early embryogenesis in vitro with pluripotent stem cells. Preprint at https://doi.org/10.1101/2021.03.12.435175 (2021).
Yu, L. et al. Blastocyst-like structures generated from human pluripotent stem cells. Nature 591, 620–626 (2021).
Liu, X. et al. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 591, 627–632 (2021).
Simunovic, M. et al. A 3D model of a human epiblast reveals BMP4-driven symmetry breaking. Nat. Cell Biol. 21, 900–910 (2019).
Moris, N. et al. An in vitro model of early anteroposterior organization during human development. Nature 582, 410–415 (2020).
Shao, Y. et al. A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat. Commun. 8, 208 (2017).
Warmflash, A., Sorre, B., Etoc, F., Siggia, E. D. & Brivanlou, A. H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 11, 847–854 (2014).
Camp, J. G., Wollny, D. & Treutlein, B. Single-cell genomics to guide human stem cell and tissue engineering. Nat. Methods 15, 661–667 (2018). This review highlights the potential utility of single-cell genomics to optimize cell and tissue engineering, with a focus on emerging methodologies that can guide this process (such as transcription factor combinatorics, spatial reconstruction, CRISPR–Cas9 screens and lineage-coupled transcriptomics).
Morgan, L. Icons of Life: A Cultural History of Human Embryos (Univ. California Press, 2009).
Blonder, L. X. Morphogenesis: The cellular and molecular processes of developmental anatomy. By Jonathan Bard. xi 313 pp. New York: Cambridge University Press. 1990. $37.95. (paper). Am. J. Hum. Biol. 5, 245–246 (1993).
Aldridge, S. & Teichmann, S. A. Single cell transcriptomics comes of age. Nat. Commun. 11, 4307 (2020).
Human Cell Atlas. Home, https://www.humancellatlas.org/ (2021).
Regev, A. et al. Science forum: the human cell atlas. eLife 6, e27041 (2017).
The HCA Consortium. The Human Cell Atlas White Paper, https://www.humancellatlas.org/wp-content/uploads/2019/11/HCA_WhitePaper_18Oct2017-copyright.pdf (2019).
Human Cell Atlas. Register, https://www.humancellatlas.org/register (2021).
Bock, C. et al. The organoid cell atlas. Nat. Biotechnol. 39, 13–17 (2021).
Subbaraman, N. Lab-grown structures mimic human embryo’s earliest stage yet. Nature 591, 510–511 (2021).
Wellcome Trust. Sharing Data from Large-scale Biological Research Projects: A System of Tripartite Responsibility, http://www.genome.gov/Pages/Research/WellcomeReport0303.pdf (Wellcome Trust, 2003).
Gerrelli, D., Lisgo, S., Copp, A. J. & Lindsay, S. Enabling research with human embryonic and fetal tissue resources. Development 142, 3073–3076 (2015). The HDBR is a biobank that collects and distributes material for research from human embryos (from 4 weeks after conception) and fetuses (up to 22 weeks after conception); https://www.hdbr.org/ shows the range of facilities offered by the HDBR and provides access for prospective users.
Human Cell Atlas. Ethics, https://www.humancellatlas.org/ethics/ (2021).
Huang, Q. et al. Intravital imaging of mouse embryos. Science 368, 181–186 (2020).
Mereu, E. et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 38, 747–755 (2020).
Argelaguet, R. et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 576, 487–491 (2019).
Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017).
McGinnis, C. S. et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat. Methods 16, 619–626 (2019).
Fujii, M., Clevers, H. & Sato, T. Modeling human digestive diseases with CRISPR-Cas9-modified organoids. Gastroenterology 156, 562–576 (2019).
Artegiani, B. et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat. Cell Biol. 22, 321–331 (2020).
Lee-Six, H. et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478 (2018).
D’Gama, A. M. & Walsh, C. A. Somatic mosaicism and neurodevelopmental disease. Nat. Neurosci. 21, 1504–1514 (2018).
Ludwig, L. S. et al. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell 176, 1325–1339 (2019).
Lareau, C. A. et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat. Biotechnol. 39, 451–461 (2021).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Ueda, H. R. et al. Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 21, 61–79 (2020).
Yang, B. et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014).
Sylwestrak, E. L., Rajasethupathy, P., Wright, M. A., Jaffe, A. & Deisseroth, K. Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell 164, 792–804 (2016).
Casoni, F. et al. Development of the neurons controlling fertility in humans: new insights from 3D imaging and transparent fetal brains. Development 143, 3969–3981 (2016).
Belle, M. et al. Tridimensional visualization and analysis of early human development. Cell 169, 161–173 (2017). A 3D map of first-trimester human development by tissue clearing and light-sheet imaging, providing high-resolution images of the developing cardiopulmonary, vascular, peripheral nervous, muscular and urogenital systems, as well as insights into complex processes such as skin innervation and the differential vascularization of male and female genital systems.
Zhao, S. et al. Cellular and molecular probing of intact human organs. Cell 180, 796–812 (2020).
Todorov, M. I. et al. Machine learning analysis of whole mouse brain vasculature. Nat. Methods 17, 442–449 (2020).
Kirst, C. et al. Mapping the fine-scale organization and plasticity of the brain vasculature. Cell 180, 780–795 (2020).
Gracia, M. et al. Mechanical impact of epithelial-mesenchymal transition on epithelial morphogenesis in Drosophila. Nat. Commun. 10, 2951 (2019).
Dumortier, J. G. et al. Hydraulic fracturing and active coarsening position the lumen of the mouse blastocyst. Science 365, 465–468 (2019).
Shahbazi, M. N., Siggia, E. D. & Zernicka-Goetz, M. Self-organization of stem cells into embryos: a window on early mammalian development. Science 364, 948–951 (2019).
Rood, J. E. et al. Toward a common coordinate framework for the human body. Cell 179, 1455–1467 (2019).
Bonneel, N. Optimal Transport for Computer Graphics and Temporal Coherence of Image Processing Algorithms. PhD thesis, Sorbonne Univ. (2018).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Popescu, D.-M. et al. Decoding human fetal liver haematopoiesis. Nature 574, 365–371 (2019). A detailed single-cell characterization of fetal liver blood and immune-cell development, revealing inferred differentiation trajectories from haematopoietic stem cells and gestation-specific potential for the differentiation of haematopoietic stem cells.
Cao, J. et al. A human cell atlas of fetal gene expression. Science 370, eaba7721 (2020). One of a set of two studies focusing on integrating single-cell gene expression55 and chromatin accessibility118 from 15 first- and second-trimester human organs.
Vento-Tormo, R. et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018). A detailed scRNA-seq analysis of first-trimester decidua and placenta, highlighting the cell–cell interactions that take place at the maternal–fetal interface during human development using a receptor–ligand database (CellPhoneDB).
Suryawanshi, H. et al. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 4, eaau4788 (2018).
Holloway, E. M. et al. Mapping development of the human intestinal niche at single-cell resolution. Cell Stem Cell 28, 568–580 (2021).
Pollen, A. A. et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 32, 1053–1058 (2014).
Han, X. et al. Construction of a human cell landscape at single-cell level. Nature 581, 303–309 (2020). A single-cell gene expression study of multiple organs during first- and second-trimester human development, with comparative analyses between human and mouse to identify conserved genetic networks.
Stewart, B. J. et al. Spatiotemporal immune zonation of the human kidney. Science 365, 1461–1466 (2019).
Cui, Y. et al. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 26, 1934–1950 (2019).
La Manno, G. et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580 (2016).
Elmentaite, R., Ross, A., James, K. R., Ortmann, D. & Gomes, T. Single-cell sequencing of developing human gut reveals transcriptional links to childhood Crohn’s disease. Dev. Cell 55, 771–783 (2020).
Young, M. D. et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599 (2018). Comparative single-cell analyses of fetal, paediatric and adult kidneys and of Wilms’ tumours, demonstrating the origin of Wilms’ tumour as aberrant nephron development.
Vértesy, Á. et al. Parental haplotype-specific single-cell transcriptomics reveal incomplete epigenetic reprogramming in human female germ cells. Nat. Commun. 9, 1873 (2018).
Li, L. et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 20, 858–873 (2017).
Nowakowski, T. J. et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017).
Camp, J. G. et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl Acad. Sci. USA 112, 15672–15677 (2015).
Lu, Y. et al. Single-cell analysis of human retina identifies evolutionarily conserved and species-specific mechanisms controlling development. Dev. Cell 53, 473–491 (2020).
Tiklová, K. et al. Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nat. Commun. 10, 581 (2019).
Kee, N. et al. Single-cell analysis reveals a close relationship between differentiating dopamine and subthalamic nucleus neuronal lineages. Cell Stem Cell 20, 29–40 (2017).
Rosenberg, A. B., Roco, C. M., Muscat, R. A. & Kuchina, A. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018).
Carter, R. A. et al. A single-cell transcriptional atlas of the developing murine cerebellum. Curr. Biol. 28, 2910–2920 (2018).
Huisman, C. et al. Single cell transcriptome analysis of developing arcuate nucleus neurons uncovers their key developmental regulators. Nat. Commun. 10, 3696 (2019).
Utz, S. G. et al. Early fate defines microglia and non-parenchymal brain macrophage development. Cell 181, 557–573 (2020).
Ginhoux, F. & Jung, S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14, 392–404 (2014).
Park, J.-E. et al. A cell atlas of human thymic development defines T cell repertoire formation. Science 367, eaay3224 (2020).
Rossant, J. & Tam, P. P. L. New insights into early human development: lessons for stem cell derivation and differentiation. Cell Stem Cell 20, 18–28 (2017).
Rayon, T. et al. Species-specific pace of development is associated with differences in protein stability. Science 369, eaba7667 (2020).
Matsuda, M. et al. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science 369, 1450–1455 (2020).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019). A densely sampled time-course analysis covering mouse gastrulation and early organogenesis provides an scRNA-seq reference atlas, which is then exploited to provide insights into early blood and endothelial development through parallel analysis of mouse chimeras that lack the key regulator TAL1 (also known as SCL).
Wagner, D. E. et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018).
Briggs, J. A. et al. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780 (2018).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Cusanovich, D. A. et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538–542 (2018).
Lescroart, F. et al. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science 359, 1177–1181 (2018).
Blakeley, P. et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 142, 3151–3165 (2015).
Deglincerti, A. et al. Self-organization of the in vitro attached human embryo. Nature 533, 251–254 (2016).
Matsuda, M. et al. Recapitulating the human segmentation clock with pluripotent stem cells. Nature 580, 124–129 (2020).
Diaz-Cuadros, M. et al. In vitro characterization of the human segmentation clock. Nature 580, 113–118 (2020).
Koike, H. et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature 574, 112–116 (2019).
Lee, J. et al. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature 582, 399–404 (2020).
Spence, J. R. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2011).
Marton, R. M. & Pașca, S. P. Organoid and assembloid technologies for investigating cellular crosstalk in human brain development and disease. Trends Cell Biol. 30, 133–143 (2020).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Quadrato, G. et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53 (2017).
Wimmer, R. A. et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 565, 505–510 (2019).
Bhaduri, A. et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature 578, 142–148 (2020).
Homsy, J. et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350, 1262–1266 (2015).
Barnat, M. et al. Huntington’s disease alters human neurodevelopment. Science 369, 787–793 (2020).
Zhang, S.-Y. et al. Human inborn errors of immunity to infection affecting cells other than leukocytes: from the immune system to the whole organism. Curr. Opin. Immunol. 59, 88–100 (2019).
Croft, B. et al. Human sex reversal is caused by duplication or deletion of core enhancers upstream of SOX9. Nat. Commun. 9, 5319 (2018).
Taylor, D. M. et al. The pediatric cell atlas: defining the growth phase of human development at single-cell resolution. Dev. Cell 49, 10–29 (2019).
Haendel, M. et al. How many rare diseases are there? Nat. Rev. Drug Discov. 19, 77–78 (2020).
Ly, A. et al. DSCAM is a netrin receptor that collaborates with DCC in mediating turning responses to netrin-1. Cell 133, 1241–1254 (2008).
Yamagishi, H. & Srivastava, D. Unraveling the genetic and developmental mysteries of 22q11 deletion syndrome. Trends Mol. Med. 9, 383–389 (2003).
Biswas, A. B. & Furniss, F. Cognitive phenotype and psychiatric disorder in 22q11.2 deletion syndrome: a review. Res. Dev. Disabil. 53-54, 242–257 (2016).
Jessa, S. et al. Stalled developmental programs at the root of pediatric brain tumors. Nat. Genet. 51, 1702–1713 (2019).
Tirosh, I. et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016).
Hovestadt, V. et al. Medulloblastomics revisited: biological and clinical insights from thousands of patients. Nat. Rev. Cancer 20, 42–56 (2020).
Phillips, H. S. et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006).
Sharma, A. et al. Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell 183, 377–394 (2020).
Reynolds, G. et al. Developmental cell programs are co-opted in inflammatory skin disease. Science 371, eaba6500 (2021). Comparative analyses of fetal skin with healthy and diseased adult skin, revealing the co-option of developmental cell programs in two common inflammatory skin conditions (atopic dermatitis and psoriasis).
Sungnak, W. et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 26, 681–687 (2020).
Barker, R. A., Parmar, M., Studer, L. & Takahashi, J. Human trials of stem cell-derived dopamine neurons for Parkinson’s disease: dawn of a new era. Cell Stem Cell 21, 569–573 (2017).
Takahashi, J. Preparing for first human trial of induced pluripotent stem cell-derived cells for Parkinson’s disease: an interview with Jun Takahashi. Regen. Med. 14, 93–95 (2019).
Domcke, S. et al. A human cell atlas of fetal chromatin accessibility. Science 370, eaba7612 (2020).
Asp, M. et al. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 179, 1647–1660 (2019). A temporal and 3D spatial map of the developing human heart from the first trimester by using a combination of transcriptome-wide scRNA-seq and spatial transcriptomics methods with cellular validation by in situ sequencing.
NHGRI & NICHD. Developmental Genotype-Tissue Expression (dGTEx). https://www.genome.gov/Funded-Programs-Projects/Developmental-Genotype-Tissue-Expression (2020).
Yan, L. et al. Single-cell RNA-seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013). A comprehensive scRNA-seq analysis of human oocytes to blastocyst-stage embryos that has widely been used to investigate lineage-associated gene expression and as a comparative analysis to human pluripotent stem cell lines.
Petropoulos, S. et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012–1026 (2016).
Tyser, R. C. V. et al. A spatially resolved single cell atlas of human gastrulation. Preprint at https://doi.org/10.1101/2020.07.21.213512 (2020).
Elmentaite, R. et al. Cells of the human intestinal tract mapped across space and time. Nature, https://doi.org/10.1038/s41586-021-03852-1 (2021).
Acknowledgements
The HDCA initiative receives funding from Wellcome, the UK Research and Innovation Medical Research Council, EU Horizon 2020, INSERM (HuDeCA) and the Knut and Alice Wallenberg and Erling-Persson foundations. We thank the HCA Executive Office and T. Andrews for their support. This publication is part of the Human Cell Atlas - https://www.humancellatlas.org/publications/
Author information
Authors and Affiliations
Consortia
Contributions
M.H., D.T. and S. Linnarsson are coordinators of the HCA Developmental Biological Network. M.H., S.A.T. and A. Regev conceived the idea, co-ordinated the writing process, wrote parts of the paper and edited all sections. A.H. designed and created the figures. All other authors wrote parts of the paper and provided feedback on all parts.
Corresponding author
Ethics declarations
Competing interests
A. Regev is a co-founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas, and was a Scientific Advisory Board member of Thermo Fisher Scientific, Syros Pharmaceuticals, Neogene Therapeutics and Asimov until 31 July 2020. From 1 August 2020, A. Regev and O.R-R. are employees of Genentech. S.A.T. has consulted for Genentech and Roche, and is a remunerated member of Scientific Advisory Boards for GlaxoSmithKline, Biogen and Foresite Labs. J. Lundeberg is a scientific advisor for 10x Genomics. All other authors declare no competing interests.
Additional information
Peer review information Nature thanks Oliver Pourquié, Susana Chuva de Sousa Lopes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Table 1
HDCA consortia listed are funded HDCA projects that have been registered with the HCA. Any researcher, including those not part of existing HDCA consortia, can contribute to HDCA.
Rights and permissions
About this article
Cite this article
Haniffa, M., Taylor, D., Linnarsson, S. et al. A roadmap for the Human Developmental Cell Atlas. Nature 597, 196–205 (2021). https://doi.org/10.1038/s41586-021-03620-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03620-1
This article is cited by
-
Decoding the spatiotemporal regulation of transcription factors during human spinal cord development
Cell Research (2024)
-
Genetic variation across and within individuals
Nature Reviews Genetics (2024)
-
Genetics of human brain development
Nature Reviews Genetics (2024)
-
Inner Ear Organoids: Strengths and Limitations
Journal of the Association for Research in Otolaryngology (2024)
-
Ethical, legal, regulatory, and policy issues concerning embryoids: a systematic review of the literature
Stem Cell Research & Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.