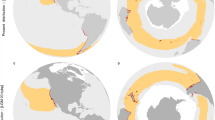
Abstract
Range shift is the primary short-term species response to rapid climate change, but it is often hampered by natural or anthropogenic habitat fragmentation. Different critical areas of a species’ niche may be exposed to heterogeneous environmental changes and modelling species response under such complex spatial and ecological scenarios presents well-known challenges. Here, we use a biophysical ecological niche model validated through population genomics and palaeodemography to reconstruct past range shifts and identify future vulnerable areas and potential refugia of the king penguin in the Southern Ocean. Integrating genomic and demographic data at the whole-species level with specific biophysical constraints, we present a refined framework for predicting the effect of climate change on species relying on spatially and ecologically distinct areas to complete their life cycle (for example, migratory animals, marine pelagic organisms and central-place foragers) and, in general, on species living in fragmented ecosystems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Thomas, C. D. et al. Extinction risk from climate change. Nature 427, 145–148 (2004).
Pacifici, M. et al. Assessing species vulnerability to climate change. Nat. Clim. Change 5, 215–224 (2015).
Charmantier, A. et al. Adaptive phenotypic plasticity in response to climate change in a wild bird population. Science 320, 800–803 (2008).
Garcia, R. A., Cabeza, M., Rahbek, C. & Araújo, M. B. Multiple dimensions of climate change and their implications for biodiversity. Science 344, 1247579 (2014).
Walther, G.-R. et al. Ecological responses to recent climate change. Nature 416, 389–395 (2002).
Gouveia, S. F. et al. Climate and land use changes will degrade the configuration of the landscape for titi monkeys in eastern Brazil. Glob. Change Biol. 22, 2003–2012 (2016).
Edwards, M. & Richardson, A. J. Impact of climate change on marine pelagic phenology and trophic mismatch. Nature 430, 881–884 (2004).
Saraux, C. et al. Reliability of flipper-banded penguins as indicators of climate change. Nature 469, 203–206 (2011).
Kearney, M. & Porter, W. Mechanistic niche modelling: combining physiological and spatial data to predict species’ ranges. Ecol. Lett. 12, 334–350 (2009).
Thuiller, W. et al. A road map for integrating eco-evolutionary processes into biodiversity models. Ecol. Lett. 16, 94–105 (2013).
Elith, J., Kearney, M. & Phillips, S. The art of modelling range‐shifting species. Methods Ecol. Evol. 1, 330–342 (2010).
Fordham, D. A. et al. Population dynamics can be more important than physiological limits for determining range shifts under climate change. Glob. Change Biol. 19, 3224–3237 (2013).
Fordham, D. A., Brook, B. W., Moritz, C. & Nogués-Bravo, D. Better forecasts of range dynamics using genetic data. Trends Ecol. Evol. 29, 436–443 (2014).
Fordham, D. A. et al. Predicting and mitigating future biodiversity loss using long-term ecological proxies. Nat. Clim. Change 6, 909–916 (2016).
Alter, S. E. et al. Climate impacts on transocean dispersal and habitat in gray whales from the Pleistocene to 2100. Mol. Ecol. 24, 1510–1522 (2015).
Kearney, M., Porter, W. P., Williams, C., Ritchie, S. & Hoffmann, A. A. Integrating biophysical models and evolutionary theory to predict climatic impacts on species’ ranges: the dengue mosquito in Australia. Funct. Ecol. 23, 528–538 (2009).
Chen, I.-C., Hill, J. K., Ohlemüller, R., Roy, D. B. & Thomas, C. D. Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024–1026 (2011).
Bost, C. A. et al. Large-scale climatic anomalies affect marine predator foraging behaviour and demography. Nat. Commun. 6, 8220 (2015).
Trucchi, E. et al. King penguin demography since the last glaciation inferred from genome-wide data. Proc. R. Soc. B 281, 20140528 (2014).
Péron, C., Weimerskirch, H. & Bost, C.-A. Projected poleward shift of king penguins’ (Aptenodytes patagonicus) foraging range at the Crozet Islands, southern Indian Ocean. Proc. R. Soc. B 279, 2515–2523 (2012).
Le Bohec, C. et al. King penguin population threatened by Southern Ocean warming. Proc. Natl Acad. Sci. USA 105, 2493–2497 (2008).
Engler, R. et al. Predicting future distributions of mountain plants under climate change: does dispersal capacity matter. Ecography 32, 34–45 (2009).
Clucas, G. V. et al. Dispersal in the sub-Antarctic: king penguins show remarkably little population genetic differentiation across their range. BMC Evol. Biol. 16, 211 (2016).
Barrat, A. Quelques aspects de la biologie et de l’écologie du manchot royal Aptenodytes patagonicus des îles Crozet. Com. Natl Fr. Rech. Antarct. 40, 9–51 (1976).
Heupink, T. H., van den Hoff, J. & Lambert, D. M. King penguin population on Macquarie Island recovers ancient DNA diversity after heavy exploitation in historic times. Biol. Lett. 8, 586–589 (2012).
Pistorius, P. A., Baylis, A., Crofts, S. & Pütz, K. Population development and historical occurrence of king penguins at the Falkland Islands. Antarct. Sci. 24, 435–440 (2012).
Kusch, A. & Marín, M. Sobre la distribución del Pingüino Rey Aptenodytes Patagonicus (Aves: Spheniscidae) en Chile. An. Inst. Patagonia 40, 157–163 (2012).
Wallberg, A. et al. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat. Genet. 46, 1081–1088 (2014).
Pearson, R. G. & Dawson, T. P. Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Glob. Ecol. Biogeog. 12, 361–371 (2003).
Bost, C.-A. et al. The importance of oceanographic fronts to marine birds and mammals of the southern oceans. J. Mar. Syst. 78, 363–376 (2009).
Wolff, E. W. et al. Southern Ocean sea-ice extent, productivity and iron flux over the past eight glacial cycles. Nature 440, 491–496 (2006).
Kohfeld, K. E. et al. Southern Hemisphere westerly wind changes during the Last Glacial Maximum: paleo-data synthesis. Quat. Sci. Rev. 68, 76–95 (2013).
Gersonde, R., Crosta, X., Abelmann, A. & Armand, L. Sea-surface temperature and sea ice distribution of the Southern Ocean at the EPILOG Last Glacial Maximum: a circum-Antarctic view based on siliceous microfossil records. Quat. Sci. Rev. 24, 869–896 (2005).
Hodgson, D. A. et al. Terrestrial and submarine evidence for the extent and timing of the Last Glacial Maximum and the onset of deglaciation on the maritime-Antarctic and sub-Antarctic islands. Quat. Sci. Rev. 100, 137–158 (2014).
Liu, X. & Fu, Y.-X. Exploring population size changes using SNP frequency spectra. Nat. Genet. 47, 555–559 (2015).
Cristofari, R. et al. Full circumpolar migration ensures evolutionary unity in the Emperor penguin. Nat. Commun. 7, 11842 (2016).
Borboroglu, P. G. & Boersma, P. D. Penguins: Natural History and Conservation (University of Washington Press, Seattle & London, 2013).
Austin, J. J. et al. The origins of the enigmatic Falkland Islands wolf. Nat. Commun. 4, 1552 (2013).
Meinshausen, M. et al. The RCP greenhouse gas concentrations and their extensions from 1765 to 2300. Clim. Change 109, 213–241 (2011).
Carr, M.-E. et al. A comparison of global estimates of marine primary production from ocean color. Deep Sea Res. II 53, 741–770 (2006).
Froneman, P. W., Laubscher, R. K. & McQuaid, C. D. Size-fractionated primary production in the south Atlantic and Atlantic sectors of the Southern Ocean. J. Plankton Res. 23, 611–622 (2001).
Pütz, K. & Cherel, Y. The diving behaviour of brooding king penguins (Aptenodytes patagonicus) from the Falkland Islands: variation in dive profiles and synchronous underwater swimming provide new insights into their foraging strategies. Mar. Biol. 147, 281–290 (2005).
Hoffmann, A. A. & Sgrò, C. M. Climate change and evolutionary adaptation. Nature 470, 479–485 (2011).
Norberg, J., Urban, M. C., Vellend, M., Klausmeier, C. A. & Loeuille, N. Eco-evolutionary responses of biodiversity to climate change. Nat. Clim. Change 2, 747–751 (2012).
Hope, A. G., Waltari, E., Payer, D. C., Cook, J. A. & Talbot, S. L. Future distribution of tundra refugia in northern Alaska. Nat. Clim. Change 3, 931–938 (2013).
Roberge, J. M. & Angelstam, P. Usefulness of the umbrella species concept as a conservation tool. Conserv. Biol. 18, 76–85 (2004).
Jackson, J. B. C. Ecological extinction and evolution in the brave new ocean. Proc. Natl Acad. Sci. USA 105, 11458–11465 (2008).
Kuhlbrodt, T. et al. An integrated assessment of changes in the thermohaline circulation. Clim. Change 96, 489–537 (2009).
Travis, J. M. Climate change and habitat destruction: a deadly anthropogenic cocktail. Proc. R. Soc. B 270, 467–473 (2003).
Ewers, R. M. & Didham, R. K. Confounding factors in the detection of species responses to habitat fragmentation. Biol. Rev. 81, 117–142 (2006).
Augustin, L. et al. Eight glacial cycles from an Antarctic ice core. Nature 429, 623–628 (2004).
Li, C. et al. Two Antarctic penguin genomes reveal insights into their evolutionary history and molecular changes related to the Antarctic environment. Gigascience 3, 27 (2014).
Zhou, Q. et al. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 346, 1246338 (2014).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Korneliussen, T. S., Albrechtsen, A. & Nielsen, R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15, 356 (2014).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Hanson-Smith, V., Kolaczkowski, B. & Thornton, J. W. Robustness of ancestral sequence reconstruction to phylogenetic uncertainty. Molecular Biol. Evol. 27, 1988–1999 (2010).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Excoffier, L., Laval, G. & Schneider, S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 (2005).
Reich, D., Thangaraj, K., Patterson, N., Price, A. L. & Singh, L. Reconstructing Indian population history. Nature 461, 489–494 (2009).
Romiguier, J. et al. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 515, 261–263 (2014).
Fumagalli, M., Vieira, F. G., Linderoth, T. & Nielsen, R. ngsTools: methods for population genetics analyses from next-generation sequencing data. Bioinformatics 30, 1486–1487 (2014).
Jombart, T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008).
Skotte, L., Korneliussen, T. S. SpringerAmpamp; Albrechtsen, A. Estimating individual admixture proportions from next generation sequencing data. Genetics 195, 693–702 (2013).
Raj, A., Stephens, M. & Pritchard, J. K. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197, 573–589 (2014).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Huson, D. H. & Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006).
Gutenkunst, R. N., Hernandez, R. D., Williamson, S. H. & Bustamante, C. D. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 (2009).
Saether, B. E. et al. Generation time and temporal scaling of bird population dynamics. Nature 436, 99–102 (2005).
Millar, C. D. et al. Mutation and evolutionary rates in Adélie penguins from the Antarctic. PLoS Genet. 4, e1000209 (2008).
Li, H. & Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011).
Schiffels, S. & Durbin, R. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46, 919–927 (2014).
Staab, P. R., Zhu, S., Metzler, D. & Lunter, G. scrm: efficiently simulating long sequences using the approximated coalescent with recombination. Bioinformatics 31, 1680–1682 (2015).
Rambaut, A. & Grass, N. C. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 13, 235–238 (1997).
Taylor, K. E., Stouffer, R. J. & Meehl, G. A. An overview of CMIP5 and the experiment design. Bull. Am. Met. Soc. 93, 485–498 (2012).
Meijers, A. J. S. The Southern Ocean in the Coupled Model Intercomparison Project phase 5. Phil. Trans. R. Soc. A 372, 20130296 (2014).
Moore, J. K., Abbott, M. R. & Richman, J. G. Location and dynamics of the Antarctic Polar Front from satellite sea surface temperature data. J. Geophys. Res. 104, 3059–3073 (1999).
Reynolds, R. W., Rayner, N. A., Smith, T. M., Stokes, D. C. & Wang, W. An improved in situ and satellite SST analysis for climate. J. Clim. 15, 1609–1625 (2002).
Adams, N. J. & Klages, N. T. Seasonal variation in the diet of the king penguin (Aptenodytes patagonicus) at sub Antarctic Marion Island. J. Zool. 212, 303–324 (1987).
Koudil, M., Charrassin, J.-B., Le Maho, Y. & Bost, C.-A. Seabirds as monitors of upper-ocean thermal structure. King penguins at the Antarctic polar front, east of Kerguelen sector. Comptes Rendus Acad. Sci. 323, 377–384 (2000).
Pütz, K. Spatial and temporal variability in the foraging areas of breeding king penguins. Condor 104, 528–538 (2002).
Moore, G. J., Robertson, G. & Wienecke, B. Food requirements of breeding king penguins at Heard Island and potential overlap with commercial fisheries. Polar Biol. 20, 293–302 (1998).
Wienecke, B. & Robertson, G. Foraging areas of king penguins from Macquarie Island in relation to a marine protected area. Environ. Manag. 29, 662–672 (2002).
Halpern, B. S. et al. A global map of human impact on marine ecosystems. Science 319, 948–952 (2008).
Turner, J., Bracegirdle, T. J., Phillips, T., Marshall, G. J. & Hosking, J. S. An initial assessment of Antarctic sea ice extent in the CMIP5 models. J. Clim. 26, 1473–1484 (2013).
Xu, S. et al. Simulation of sea ice in FGOALS-g2: Climatology and late 20th century changes. Adv. Atmos. Sci. 30, 658–673 (2013).
Shu, Q., Song, Z. & Qiao, F. Assessment of sea ice simulations in the CMIP5 models. Cryosphere 9, 399–409 (2015).
Goberville, E., Beaugrand, G., Hautekèete, N. C., Piquot, Y. & Luczak, C. Uncertainties in the projection of species distributions related to general circulation models. Ecol. Evol. 5, 1100–1116 (2015).
Raybaud, V. et al. Decline in kelp in west Europe and climate. PloS One 8, e66044 (2013).
Cabré, A., Marinov, I., Bernardello, R. & Bianchi, D. Oxygen minimum zones in the tropical Pacific across CMIP5 models: mean state differences and climate change trends. Biogeosciences 12, 5429–5454 (2015).
Acknowledgements
This work was conducted within the framework of the Programme 137 of the Institut Polaire Français Paul-Emile Victor (IPEV; CLB), with additional support from the French National Research Agency (ANR) ‘PICASO’ grant (ANR-2010-BLAN-1728-01; Y.L.M.), Marie Curie Intra European Fellowships (FP7-PEOPLE-IEF-2008, European Commission; project no. 235962 to C.L.B. and FP7-PEOPLE-IEF-2010, European Commission; project no. 252252 to E.T.), the Centre Scientifique de Monaco through the budget allocated to the Laboratoire International Associé 647 BioSensib (CSM/CNRS-University of Strasbourg; C.L.B., Y.L.M.), the Centre National de la Recherche Scientifique (Programme Zone Atelier de Recherches sur l’Environnement Antarctique et Subantarctique), South African National Antarctic Programme (P.P.) and the IPEV Programme 109 (Y.C.). Logistic and field costs of research were supported by the IPEV Programme 137 (C.L.B.), the South African Department of Environmental Affairs and the National Research Foundation (P.P.). This work was performed on the Abel Cluster, owned by the University of Oslo and the Norwegian Metacenter for High Performance Computing (NOTUR), and operated by the Department for Research Computing at USIT, the University of Oslo. We are very grateful to M. Skage, A. Tooming-Klunderud, M. Selander-Hansen and the Norwegian Sequencing Center for their very valuable help in the laboratory, as well as L. Nederbragt and M. Matschiner for their assistance with the Abel cluster, and M. Fumagalli and T. Korneliussen for their precious advice regarding ngsTools and ANGSD. We thank G. Bertorelle, L. Fusani, A. Mazzarella and D. Fordham for useful comments and advice. We acknowledge the World Climate Research Programme’s Working Group on Coupled Modelling, which is responsible for CMIP, and we thank the climate modelling groups (listed in Supplementary Table 1) for producing and making available their model output. For CMIP, the US Department of Energy’s Program for Climate Model Diagnosis and Intercomparison provides coordinating support and led development of software infrastructure in partnership with the Global Organization for Earth System Science Portals.
Author information
Authors and Affiliations
Contributions
C.L.B. and E.T. conceived and supervised the study. C.L.B., F.B., Y.C. and P.P. collected the samples. R.C. performed DNA extraction, library preparation, and prepared and performed the genomic and demographic analyses and the climate modelling. X.L. and E.T. participated in the genomic and demographic analyses. V.R. and C.L.B. participated in climate modelling. N.C.S. hosted the project. R.C., C.L.B. and E.T. wrote the manuscript. F.B., N.C.S., P.P., Y.C., Y.L.M. and V.R. commented the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–5, Supplementary Figures 1–9, Supplementary Tables 1–4, Supplementary References
Rights and permissions
About this article
Cite this article
Cristofari, R., Liu, X., Bonadonna, F. et al. Climate-driven range shifts of the king penguin in a fragmented ecosystem. Nature Clim Change 8, 245–251 (2018). https://doi.org/10.1038/s41558-018-0084-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41558-018-0084-2
This article is cited by
-
Past volcanic activity predisposes an endemic threatened seabird to negative anthropogenic impacts
Scientific Reports (2024)
-
Distribution shifts in Indo-Pacific humpback dolphins and the co-occurrence of marine heatwaves
Reviews in Fish Biology and Fisheries (2024)
-
Genome-wide assessment of population genetic and demographic history in Magnolia odoratissima based on SLAF-seq
Conservation Genetics (2023)
-
The global ecological niche of lumpfish (Cyclopterus lumpus) and predicted range shifts under climate change
Hydrobiologia (2023)
-
Records of king penguins at Stranger Point and Esperanza/Hope Bay, Antarctica
Polar Biology (2023)