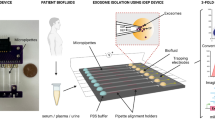
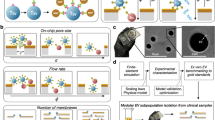
Abstract
Extracellular vesicles (EVs) can mediate intercellular communication by transferring cargo proteins and nucleic acids between cells. The pathophysiological roles and clinical value of EVs are under intense investigation, yet most studies are limited by technical challenges in the isolation of nanoscale EVs (nEVs). Here, we report a lipid-nanoprobe system that enables spontaneous labelling of nEVs for subsequent magnetic enrichment in 15 minutes, with isolation efficiency and cargo composition similar to what can be achieved by the much slower and bulkier method of ultracentrifugation. We also show that this approach allows for downstream analyses of nucleic acids and proteins, enabling the identification of EGFR and KRAS mutations following nEV isolation from the blood plasma of non-small-cell lung-cancer patients. The efficiency and versatility of the lipid-nanoprobe approach opens up opportunities in point-of-care cancer diagnostics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Colombo, M., Raposo, G. & Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell. Dev. Biol. 30, 255–289 (2014).
Raposo, G. & Stoorvogel, W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 (2013).
Cocucci, E. & Meldolesi, J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 25, 364–372 (2015).
Yáñez-Mó, M. et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066 (2015).
Thery, C., Zitvogel, L. & Amigorena, S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 (2002).
Li, Y., Shen, Z. & Yu, X.-Y. Transport of microRNAs via exosomes. Nat. Rev. Cardiol. 12, 198–198 (2015).
Alderton, G. K. Diagnosis: fishing for exosomes. Nat. Rev. Cancer 15, 453–453 (2015).
Melo, S. A. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523, 177–182 (2015).
Thakur, B. K. et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 24, 766–769 (2014).
Kahlert, C. et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 289, 3869–3875 (2014).
Costa-Silva, B. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816–826 (2015).
Melo, S. A. et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721 (2014).
Yeo, R. W. Y. et al. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv. Drug Deliv. Rev. 65, 336–341 (2013).
Lai, R. C., Yeo, R. W. Y., Tan, K. H. & Lim, S. K. Exosomes for drug delivery—a novel application for the mesenchymal stem cell. Biotechnol. Adv. 31, 543–551 (2013).
Azmi, A. S., Bao, B. & Sarkar, F. H. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metast. Rev. 32, 623–642 (2013).
Ohno, S.-i. et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 21, 185–191 (2013).
Liga, A., Vliegenthart, A. D. B., Oosthuyzen, W., Dear, J. W. & Kersaudy-Kerhoas, M. Exosome isolation: a microfluidic road-map. Lab Chip 15, 2388–2394 (2015).
Christianson, H. C., Svensson, K. J., van Kuppevelt, T. H., Li, J.-P. & Belting, M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl Acad. Sci. USA 110, 17380–17385 (2013).
Bechstein, D. J. B. et al. High performance wash-free magnetic bioassays through microfluidically enhanced particle specificity. Sci. Rep. 5, 11693 (2015).
Caballero, J. N., Frenette, G., Belleannée, C. & Sullivan, R. CD9-positive microvesicles mediate the transfer of molecules to bovine spermatozoa during epididymal maturation. PLoS ONE 8, e65364 (2013).
Tauro, B. J. et al. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell. Proteomics 12, 587–598 (2013).
Thery, C. et al. Indirect activation of naive CD4+ T cells by dendritic cell-derived exosomes. Nat. Immunol. 3, 1156–1162 (2002).
Lambertz, U. et al. Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world Leishmania providing evidence for conserved exosomal RNA packaging. BMC Genomics 16, 1–26 (2015).
Gormally, E., Caboux, E., Vineis, P. & Hainaut, P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat. Res. 635, 105–117 (2007).
Wei, Z., Batagov, A. O., Carter, D. R. & Krichevsky, A. M. Fetal bovine serum RNA interferes with the cell culture derived extracellular RNA. Sci. Rep. 6, 31175 (2016).
Kowal, J. et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl Acad. Sci. USA 113, E968–E977 (2016).
Asano, H. et al. Detection of EGFR gene mutation in lung cancer by mutant-enriched polymerase chain reaction assay. Clin. Cancer Res. 12, 43–48 (2006).
Vlassov, A. V., Magdaleno, S., Setterquist, R. & Conrad, R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 1820, 940–948 (2012).
Klymchenko, A. S. & Kreder, R. Fluorescent probes for lipid rafts: from model membranes to living cells. Chem. Biol. 21, 97–113 (2014).
Wijesinghe, D., Arachchige, M. C. M., Lu, A., Reshetnyak, Y. K. & Andreev, O. A. pH dependent transfer of nano-pores into membrane of cancer cells to induce apoptosis. Sci. Rep. 3, 3560 (2013).
Lobb, R. J. et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 4, 27031 (2015).
Jeong, S. et al. Integrated magneto–electrochemical sensor for exosome analysis. ACS Nano 10, 1802–1809 (2016).
Im, H. et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 32, 490–495 (2014).
Zhao, Z., Yang, Y., Zeng, Y. & He, M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 16, 489–496 (2016).
Weber, R. J., Liang, S. I., Selden, N. S., Desai, T. A. & Gartner, Z. J. Efficient targeting of fatty-acid modified oligonucleotides to live cell membranes through stepwise assembly. Biomacromolecules 15, 4621–4626 (2014).
Charbonneau, D. M. & Tajmir-Riahi, H.-A. Study on the interaction of cationic lipids with bovine serum albumin. J. Phys. Chem. B 114, 1148–1155 (2010).
Bobrie, A., Colombo, M., Krumeich, S., Raposo, G. & Théry, C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles 1, 18397 (2012).
Van Niel, G. et al. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 121, 337–349 (2001).
Colombo, M. et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 126, 5553–5565 (2013).
Rupp, A. K. et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol. Oncol. 122, 437–446 (2011).
Boch, C. et al. The frequency of EGFR and KRAS mutations in non-small cell lung cancer (NSCLC): routine screening data for central Europe from a cohort study. BMJ Open 3, e002560 (2013).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies . Sci. Transl. Med. 6, 224ra224 (2014).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Cheng, G. et al. The GO/rGO-Fe3O4 composites with good water-dispersibility and fast magnetic response for effective immobilization and enrichment of biomolecules. J. Mater. Chem. 22, 21998–22004 (2012).
Cheng, G., Zhang, J.-L., Liu, Y.-L., Sun, D.-H. & Ni, J.-Z. Synthesis of novel Fe3O4@SiO2@CeO2 microspheres with mesoporous shell for phosphopeptide capturing and labeling. Chem. Comm. 47, 5732–5734 (2011).
Deng, Y., Qi, D., Deng, C., Zhang, X. & Zhao, D. Superparamagnetic high-magnetization microspheres with an Fe3O4@ SiO2 core and perpendicularly aligned mesoporous SiO2 shell for removal of microcystins. J. Am. Chem. Soc. 130, 28–29 (2008).
Morel, A.-L. et al. Sonochemical approach to the synthesis of Fe3O4@SiO2 core−shell nanoparticles with tunable properties. ACS Nano 2, 847–856 (2008).
Ding, H. et al. Fe3O4@SiO2 core/shell nanoparticles: the silica coating regulations with a single core for different core sizes and shell thicknesses. Chem. Mater. 24, 4572–4580 (2012).
Wan, Y. et al. Surface-immobilized aptamers for cancer cell isolation and microscopic cytology. Cancer Res. 70, 9371–9380 (2010).
Wang, S., Wan, Y. & Liu, Y. Effects of nanopillar array diameter and spacing on cancer cell capture and cell behaviors. Nanoscale 6, 12482–12489 (2014).
Xue, P. et al. Isolation and elution of Hep3B circulating tumor cells using a dual-functional herringbone chip. Microfluid. Nanofluid. 16, 605–612 (2014).
Lu, N.-N . et al. Biotin-triggered decomposable immunomagnetic beads for capture and release of circulating tumor cells. ACS Appl. Mater. Interfaces 7, 8817–8826 (2015).
Wan, Y. et al. Nanotextured substrates with immobilized aptamers for cancer cell isolation and cytology. Cancer 118, 1145–1154 (2012).
Lynch, T. J. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004).
Li, H & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25, 1754–17320 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Dobin, A . et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Karolchik, D., Hinrichs, A. S. & Kent, W. J. The UCSC Genome Browser. Curr. Protoc. Bioinform. Ch. 1, Unit 1 4 (2007).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Alam, S. et al. The eleventh and twelfth data releases of the Sloan Digital Sky Survey: final data from SDSS-III. Astrophys. J. Suppl. Ser. 219, 12 (2015).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R016 (2010).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
Wan, Y. et al. Dataset for rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes. figsharehttp://dx.doi.org/10.6084/m9.figshare.4728856 (2017).
Acknowledgements
S.-Y.Z. thanks the Penn State Materials Research Institute, the Huck Institute of Life Sciences, the Penn State Hershey Cancer Institute, the Penn State proteomic and mass spectrometry facilities at Hershey and University Park, the Penn State Microscopy and Cytometry Facility, and the Penn State Genomics Facility for their support. This work was partially supported by the Pennsylvania State University start-up fund and the National Cancer Institute of the National Institutes of Health under Award Number DP2CA174508. We thank the Applied Bioinformatics Center (BFX) at the New York University (NYU) School of Medicine for providing bioinformatics support and for helping with the analysis and interpretation of the data. This work used computing resources at the High Performance Computing Facility (HPCF) of the Center for Health Informatics and Bioinformatics at the NYU Langone Medical Center. We also thank the Genome Technology Center (GTC) for library preparation and sequencing. This shared resource is partially supported by the Cancer Center Support Grant, P30CA016087, at the Laura and Isaac Perlmutter Cancer Center. We would like to thank S. Hafenstein at Penn State Hershey for discussions on cryo-SEM and C. Zhang at the Dana-Farber Cancer Institute for his advice on genomic analysis.
Author information
Authors and Affiliations
Contributions
Y.W. and S.-Y.Z. designed the research. Y.W. conducted experiments and analysed data. G.C. prepared the MMPs, assisted with peptide-sample preparation and performed proteomic analyses. S.-J.H. assisted with the preparation of NGS samples, the analysis of RNA NGS data, and the fluorescence imaging. M.N. prepared blood plasma. C.-D.Z. and W.-Q.L. assisted with the cell culture, nEV collection and gel electrophoresis. Y.-Q.X. performed the wound-healing assay. Z.-G.W. performed the electron microscopy. W.-L.Z. assisted with the image processing. A.S and I.A analysed NGS DNA data. X.L., S.J.R. and C.P.B. recruited patients and provided blood samples, tissue NGS data and clinical support. Y.W. and S.-Y.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary tables, figures and references (PDF 7817 kb)
Supplementary Dataset 1
Top-1,000 expressed mRNAs (XLSX 47 kb)
Supplementary Dataset 2
Top-1,000 expressed miRNAs (XLSX 43 kb)
Supplementary Dataset 3
Cargo proteins in the nanoscale extracellular vesicles (XLSX 384 kb)
Rights and permissions
About this article
Cite this article
Wan, Y., Cheng, G., Liu, X. et al. Rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes. Nat Biomed Eng 1, 0058 (2017). https://doi.org/10.1038/s41551-017-0058
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41551-017-0058
This article is cited by
-
Exploiting sound for emerging applications of extracellular vesicles
Nano Research (2024)
-
Extracellular vesicle-based liquid biopsy biomarkers and their application in precision immuno-oncology
Biomarker Research (2023)
-
Clinical application and detection techniques of liquid biopsy in gastric cancer
Molecular Cancer (2023)
-
Recent advances of small extracellular vesicle biomarkers in breast cancer diagnosis and prognosis
Molecular Cancer (2023)
-
Exosomes as a new frontier of cancer liquid biopsy
Molecular Cancer (2022)