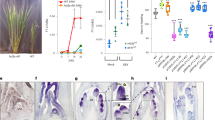
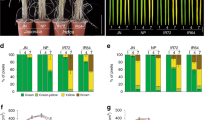
Abstract
For genetically homogeneous crops, the timing of flowering is determined largely by the cultivation environment and is strongly associated with the yield and quality of the harvest1. Flowering time and other agronomical traits are often tightly correlated, which can lead to difficulty excluding the effects of flowering time when evaluating the characteristics of different genetic varieties2. Here, we describe the development of transgenic rice plants whose flowering time can be controlled by specific agrochemicals. We first developed non-flowering rice plants by overexpressing a floral repressor gene, Grain number, plant height and heading date 7 (Ghd7)3,4, to inhibit any environmentally induced spontaneous flowering. We then co-transformed plants with a rice florigen gene, Heading date 3a (Hd3a)5, which is induced by the application of specific agrochemicals. This permitted the flowering time to be experimentally controlled regardless of the cultivation environment: some transgenic plants flowered only after agrochemical treatment. Furthermore, plant size and yield-related traits could, in some cases, be increased owing to both a longer duration of vegetative growth and an increased panicle size. This ability to control flowering time experimentally, independently of environmental variables, may lead to production of crops suitable for growth in different climates and facilitate breeding for various agronomical traits.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jung, C. & Müller, A. E. Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573 (2009).
Yamamoto, T., Yonemaru, J. & Yano, M. Towards the understanding of complex traits in rice: substantially or superficially? DNA Res. 16, 141–154 (2009).
Xue, W. et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767 (2008).
Weng, X. et al. Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 164, 735–747 (2014).
Doi, K. et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936 (2004).
Kojima, S. et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105 (2002).
Ogiso-Tanaka, E. et al. Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PLoS ONE 8, e75959 (2013).
Komiya, R., Yokoi, S. & Shimamoto, K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136, 3443–3450 (2009).
Itoh, H., Nonoue, Y., Yano, M. & Izawa, T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42, 635–638 (2010).
Itoh, H. & Izawa, T. The coincidence of critical day length recognition for florigen gene expression and floral transition under long-day conditions in rice. Mol. Plant 6, 635–649 (2013).
Hung, H. Y. et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl Acad. Sci. USA 109, E1913–E1921 (2012).
Yang, S. et al. Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12. PLoS ONE 9, e105352 (2014).
Komiya, R., Ikegami, A., Tamaki, S., Yokoi, S. & Shimamoto, K. Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774 (2008).
Matsubara, K. et al. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J. 66, 603–612 (2011).
Hori, K. et al. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 76, 36–46 (2013).
Oikawa, T. & Kyozuka, J. Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21, 1095–1108 (2009).
Miyoshi, K. et al. PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc. Natl Acad. Sci. USA 101, 875–880 (2004).
Nemoto, Y., Nonoue, Y., Yano, M. & Izawa, T. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 86, 221–233 (2016).
Ito, Y. & Kurata, N. Disruption of KNOX gene suppression in leaf by introducing its cDNA in rice. Plant Sci. 174, 357–365 (2008).
Shimono, M. et al. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076 (2007).
Umemura, K. et al. Contribution of salicylic acid glucosyltransferase, OsSGT1, to chemically induced disease resistance in rice plants. Plant J. 57, 463–472 (2009).
Günl, M., Liew, E. F., David, K. & Putterill, J. Analysis of a post-translational steroid induction system for GIGANTEA in Arabidopsis. BMC Plant Biol. 9, 141 (2009).
Krzymuski, M. et al. The dynamics of FLOWERING LOCUS T expression encodes long-day information. Plant J. 83, 952–961 (2015).
Yeoh, C. C., Balcerowicz, M., Laurie, R., Macknight, R. & Putterill, J. Developing a method for customized induction of flowering. BMC Biotechnol. 11, 36 (2011).
Saijo, T. & Nagasawa, A. Development of a tightly regulated and highly responsive copper-inducible gene expression system and its application to control of flowering time. Plant Cell Rep. 33, 47–59 (2014).
Zhang, H. et al. Precocious flowering in trees: the FLOWERING LOCUS T gene as a research and breeding tool in Populus. J. Exp. Bot. 61, 2549–2560 (2010).
Kim, S. L., Choi, M., Jung, K. H. & An, G. Analysis of the early-flowering mechanisms and generation of T-DNA tagging lines in Kitaake, a model rice cultivar. J. Exp. Bot. 64, 4169–4182 (2013).
Izawa, T. et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 23, 1741–1755 (2011).
Breitling, R., Armengaud, P., Amtmann, A. & Herzyk, P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573, 83–92 (2004).
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S. & Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036 (2007).
Endo-Higashi, N. & Izawa, T. Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice. Plant Cell Physiol. 52, 1083–1094 (2011).
Sugio, T., Satoh, J., Matsuura, H., Shinmyo, A. & Kato, K. The 5′-untranslated region of the Oryza sativa alcohol dehydrogenase gene functions as a translational enhancer in monocotyledonous plant cells. J. Biosci. Bioeng. 105, 300–302 (2008).
Ochman, H., Gerber, A. S. & Hartl, D. L. Genetic applications of an inverse polymerase chain reaction. Genetics 120, 621–623 (1988).
Taoka, K. et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335 (2011).
Abe, M. et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056 (2005).
Acknowledgements
This work was supported by grants from MAFF, Japan (Genomics for Agricultural Innovation, GPN-1001; Genomics-based Technology for Agricultural Improvement, GMO-1005; PFT-1001) to T.I.
Author information
Authors and Affiliations
Contributions
T.I. conceived and conducted the entire experiments of this work. R.O. performed most of experiments in this work. Y.N. performed the analysis of Ghd7-overexpressing plant in rice. N.E.-H. performed the effect of Hd3a ectopic expression in the Ghd7-overexpressing plant in rice. T.I. and R.O. wrote the manuscript and the others revised it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–10. (PDF 1192 kb)
Rights and permissions
About this article
Cite this article
Okada, R., Nemoto, Y., Endo-Higashi, N. et al. Synthetic control of flowering in rice independent of the cultivation environment. Nature Plants 3, 17039 (2017). https://doi.org/10.1038/nplants.2017.39
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2017.39
This article is cited by
-
Exploring Flowering Genes in Isabgol (Plantago ovata Forsk.) Through Transcriptome Analysis
Plant Molecular Biology Reporter (2021)
-
Automatic estimation of heading date of paddy rice using deep learning
Plant Methods (2019)
-
Lessons from natural variations: artificially induced heading date variations for improvement of regional adaptation in rice
Theoretical and Applied Genetics (2019)
-
Flowering time regulation: Agrochemical control of flowering
Nature Plants (2017)
-
Saving the world
Nature Plants (2017)