Abstract
Regeneration of CNS myelin involves differentiation of oligodendrocytes from oligodendrocyte progenitor cells. In multiple sclerosis, remyelination can fail despite abundant oligodendrocyte progenitor cells, suggesting impairment of oligodendrocyte differentiation. T cells infiltrate the CNS in multiple sclerosis, yet little is known about T cell functions in remyelination. We report that regulatory T cells (Treg) promote oligodendrocyte differentiation and (re)myelination. Treg-deficient mice exhibited substantially impaired remyelination and oligodendrocyte differentiation, which was rescued by adoptive transfer of Treg. In brain slice cultures, Treg accelerated developmental myelination and remyelination, even in the absence of overt inflammation. Treg directly promoted oligodendrocyte progenitor cell differentiation and myelination in vitro. We identified CCN3 as a Treg-derived mediator of oligodendrocyte differentiation and myelination in vitro. These findings reveal a new regenerative function of Treg in the CNS, distinct from immunomodulation. Although the cells were originally named 'Treg' to reflect immunoregulatory roles, this also captures emerging, regenerative Treg functions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fünfschilling, U. et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–521 (2012).
Lee, Y. et al. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature 487, 443–448 (2012).
Miron, V.E. et al. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat. Neurosci. 16, 1211–1218 (2013).
Ruckh, J.M. et al. Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell 10, 96–103 (2012).
Bieber, A.J., Kerr, S. & Rodriguez, M. Efficient central nervous system remyelination requires T cells. Ann. Neurol. 53, 680–684 (2003).
Woodruff, R.H., Fruttiger, M., Richardson, W.D. & Franklin, R.J.M. Platelet-derived growth factor regulates oligodendrocyte progenitor numbers in adult CNS and their response following CNS demyelination. Mol. Cell. Neurosci. 25, 252–262 (2004).
Arnett, H.A. et al. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science 306, 2111–2115 (2004).
Ousman, S.S. & David, S. Lysophosphatidylcholine induces rapid recruitment and activation of macrophages in the adult mouse spinal cord. Glia 30, 92–104 (2000).
Burzyn, D. et al. A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295 (2013).
Arpaia, N. et al. A distinct function of regulatory T Cells in tissue protection. Cell 162, 1078–1089 (2015).
Kim, J.M., Rasmussen, J.P. & Rudensky, A.Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197 (2007).
Christiaansen, A.F., Boggiatto, P.M. & Varga, S.M. Limitations of Foxp3(+) Treg depletion following viral infection in DEREG mice. J. Immunol. Methods 406, 58–65 (2014).
Meyer Zu Hörste, G. et al. Active immunization induces toxicity of diphtheria toxin in diphtheria resistant mice--implications for neuroinflammatory models. J. Immunol. Methods 354, 80–84 (2010).
Mayer, C.T. et al. Advantages of Foxp3(+) regulatory T cell depletion using DEREG mice. Immun. Inflamm. Dis. 2, 162–165 (2014).
Huang, J.K. et al. Retinoid X receptor gamma signaling accelerates CNS remyelination. Nat. Neurosci. 14, 45–53 (2011).
Zhang, H., Jarjour, A.A., Boyd, A. & Williams, A. Central nervous system remyelination in culture--a tool for multiple sclerosis research. Exp. Neurol. 230, 138–148 (2011).
Hild, W. Myelinbildung in Kulturen des Zentralnervensystems. Verh. Anat. Ges. 53, 315–317 (1956-1957).
Birgbauer, E., Rao, T.S. & Webb, M. Lysolecithin induces demyelination in vitro in a cerebellar slice culture system. J. Neurosci. Res. 78, 157–166 (2004).
Ling, C., Verbny, Y.I., Banks, M.I., Sandor, M. & Fabry, Z. In situ activation of antigen-specific CD8+ T cells in the presence of antigen in organotypic brain slices. J. Immunol. 180, 8393–8399 (2008).
Kim, J. et al. PINK1 deficiency enhances inflammatory cytokine release from acutely prepared brain slices. Exp. Neurobiol. 22, 38–44 (2013).
Sheridan, G.K. & Dev, K.K. S1P1 receptor subtype inhibits demyelination and regulates chemokine release in cerebellar slice cultures. Glia 60, 382–392 (2012).
Su, B.Y. et al. The expression of CCN3 (nov) RNA and protein in the rat central nervous system is developmentally regulated. Mol. Pathol. 54, 184–191 (2001).
Leask, A. & Abraham, D.J. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J. Cell Sci. 119, 4803–4810 (2006).
Lin, C.G. et al. CCN3 (NOV) is a novel angiogenic regulator of the CCN protein family. J. Biol. Chem. 278, 24200–24208 (2003).
Wang, X., He, H., Wu, X., Hu, J. & Tan, Y. Promotion of dentin regeneration via CCN3 modulation on Notch and BMP signaling pathways. Biomaterials 35, 2720–2729 (2014).
Kotter, M.R., Setzu, A., Sim, F.J., Van Rooijen, N. & Franklin, R.J.M. Macrophage depletion impairs oligodendrocyte remyelination following lysolecithin-induced demyelination. Glia 35, 204–212 (2001).
Perbal, B. NOV (nephroblastoma overexpressed) and the CCN family of genes: structural and functional issues. Mol. Pathol. 54, 57–79 (2001).
Perbal, B. CCN proteins: a centralized communication network. J. Cell Commun. Signal. 7, 169–177 (2013).
Le Dréau, G. et al. NOV/CCN3 promotes maturation of cerebellar granule neuron precursors. Mol. Cell. Neurosci. 43, 60–71 (2010).
Xin, L.W., Martinerie, C., Zumkeller, W., Westphal, M. & Perbal, B. Differential expression of novH and CTGF in human glioma cell lines. Clin. Mol. Pathol. 49, M91–M97 (1996).
Raposo, C. et al. CNS repair requires both effector and regulatory T cells with distinct temporal and spatial profiles. J. Neurosci. 34, 10141–10155 (2014).
Saxena, A. et al. Regulatory T cells are recruited in the infarcted mouse myocardium and may modulate fibroblast phenotype and function. Am. J. Physiol. Heart Circ. Physiol. 307, H1233–H1242 (2014).
Weirather, J. et al. Foxp3+ CD4+ T cells improve healing after myocardial infarction by modulating monocyte/macrophage differentiation. Circ. Res. 115, 55–67 (2014).
Wang, Y. et al. Th2 lymphoproliferative disorder of LatY136F mutant mice unfolds independently of TCR-MHC engagement and is insensitive to the action of Foxp3+ regulatory T cells. J. Immunol. 180, 1565–1575 (2008).
Hall, S.M. The effect of injections of lysophosphatidyl choline into white matter of the adult mouse spinal cord. J. Cell Sci. 10, 535–546 (1972).
Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Liu, J. et al. Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat. Neurosci. 15, 1621–1623 (2012).
Zhao, C., Li, W.W. & Franklin, R.J. Differences in the early inflammatory responses to toxin-induced demyelination are associated with the age-related decline in CNS remyelination. Neurobiol. Aging 27, 1298–1307 (2006).
Mei, F. et al. Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nat. Med. 20, 954–960 (2014).
Chevalier, G. et al. novH: differential expression in developing kidney and Wilm?s tumors. Am. J. Pathol. 152, 1563–1575 (1998).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
We thank A. Rudensky (Memorial Sloan Kettering Cancer Centre) for providing Foxp3-DTR mice and B. Malissen (Aix Marseille Université) for the provision of Foxp3-eGFP mice. We acknowledge extensive technical support from S. Leech, S. Peoples, N. de la Vega Gallardo, S. Mitchell, J. Brown, R. Blain and the staff of the animal facility. This work was supported by the Biotechnology and Biological Sciences Research Council (BB/J01026X/1 and BB/N003721/1, to D.C.F.), The Leverhulme Trust (ECF-2014-390, to Y.D.), QUB (QUB - Lucy McGuigan Bequest, to D.C.F.), The UK Multiple Sclerosis Society (941 and 50, to R.J.M.F. and C.Z.), MRC UK Regenerative Medicine platform (MR/KO26666/1, to A.C.W.), University of Edinburgh Wellcome Trust Multi User Equipment Grant (WT104915MA, to A.C.W.), by a core support grant from the Wellcome Trust and MRC to the Wellcome Trust - Medical Research Council Cambridge Stem Cell Institute (097922/Z/11/Z to R.J.M.F.), studentship support from Dept. for the Economy (Northern Ireland) and British Pathological Society, US National Multiple Sclerosis Society (RG5203A4, to J.R.C.), NIH/NINDS (NS095889, to J.R.C.), NIH/NIGMS IRACDA Postdoctoral Fellowship (K12GM081266, to S.R.M.) and Wellcome Trust (110138/Z/15/Z, to D.C.F.).
Author information
Authors and Affiliations
Contributions
Experiments were designed, performed and analyzed by Y.D., T.O.H., M.D., R.P., S.R.M., S.F., M.N., G.E., J.M., J.F., I.V.A., J.R.C. and D.C.F. Image analysis tools were developed by P.B. and P.H. EM was performed and analyzed by E.E., A.B. and A.C.W. C.Z., R.H., A.K., P.N.M., B.P., R.J.I., J.R.C. and R.J.M.F. provided advice on experimental design and interpretation, and I.V.A., P.M. and R.J.M.F. provided mentorship. Manuscript was written by Y.D. and D.C.F. with contributions from all authors. D.C.F. oversaw the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Immunophenotyping and oligodendroglial analysis following lysolecithin- and cuprizone-induced demyelination
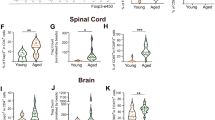
(a-d) Representative images of flow cytometric analysis of (a) CD3+ T cells, (b) CD4+ T cells and (c,d) CD3+CD4+Foxp3+ Treg, in lesional spinal cord tissue at 3 d.p.l. gated on (a,b) CD45+ cells, (c) CD45+CD3+ and (d) CD45+CD3+CD4+ cells. (e) Quantitative immunophenotyping of T cell populations in matched healthy and lesioned spinal cord harvested at 3 and 11 d.p.l., gated on CD45+ cells. n = 4 mice per group. Healthy vs. lesion on 3 d.p.l. on left panel: (CD3+, U = 0, P = 0.0286), (CD4+, U = 0, P = 0.0286), (CD4+Foxp3+, U = 0, P = 0.0294). Healthy vs. lesion on 11 d.p.l. on right panel: (CD3+, U = 2, P = 0.1143), (CD4+, U = 0, P = 0.0286), (CD4+Foxp3+, U = 0, P = 0.0294), all two-tailed Mann Whitney U tests. (f) Representative flow cytometric analysis of Foxp3+ Treg cells in fresh spleen from DT-treated C57BL/6 (control) and Foxp3-DTR mice (- Treg) at 14 d.p.l., gated on CD4+ cells. (g) Olig2+Ki67+ cells per lesion area in spinal cord of Foxp3-DTR mice at 10 d.p.l. with n = 8 animals in control and n = 7 animals in Treg-depleted groups. (t = 1.580, d.f. = 13, P = 0.1382, unpaired two-tailed Student’s t test). (h) CC1+Olig2+ cells per lesion area at 10 d.p.l. in spinal cord of mice, with Treg depletion restricted to pre-lesioning phase, with n=3 animals in control and n=4 animals in Treg-depleted groups. (U = 6, P > 0.9999, two-tailed Mann Whitney U test). (i) Immunohistochemical analysis of CC1+Olig2+ cells per lesion area of the corpus callosum at 10 days post-cuprizone withdrawal. n = 5 animals for C57BL/6 DT-treated controls, n = 4 animals for saline-treated Foxp3-DTR controls, n = 5 animals for Treg-depleted Foxp3-DTR group; data represent analysis of 2-4 regions of corpus callosum per animal (C57BL/6+DT vs FoxP3-DTR+DT: t = 0.8753, d.f. = 8, P = 0.4069, unpaired two-tailed Student’s t test; FoxP3-DTR+saline vs FoxP3-DTR+DT: U = 6, P = 0.4127, two-tailed Mann Whitney U test). (j) Plp1 expression analysis in cerebellar tissue of control and Treg-depleted animals at 14 days post-cuprizone withdrawal by qPCR (left panel, normalized to Gapdh) and by in situ hybridization in corpus callosum (right panel, representative images). n = 5 animals (control) and n = 6 animals in Treg-depleted group (t = 8.684, d.f. = 9, P < 0.0001, unpaired two-tailed Student’s t test). (k) Immunohistochemical analysis of total Olig2+ cells per lesion area in the corpus callosum at 10 and 14 days post-cuprizone withdrawal. n = 6 animals (control), n = 4 animals (demyelination and 10 days control remyelination group), n=5 (all other remyelination groups); data represent analysis of 2-4 regions of corpus callosum per animal (10 days remyelination: U = 5, P = 0.2857, two-tailed Mann Whitney U test; 14 days remyelination: t = 1.701, d.f. = 8, P = 0.1273, unpaired two-tailed Student’s t test). Data shown are representative of at least 3 (a-d, f), 2 (e, left panel) or 1 (e, right panel, g-k) independent experiments. Data presented with mean values indicated, error bars = SEM, *P<0.05, ***P<0.001.
Supplementary Figure 2 Treg and Treg-conditioned media promote brain slice myelination and remyelination
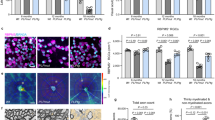
(a) Schematic diagram and 3D image of Foxp3-eGFP+ Treg in organotypic brain slice cultures representing z-stack images at 3 d.i.v. (scale bar = 100 μm, green = eGFP). (b,c) Analysis of (b) MBP+ cells per FOV and (c) myelination index (NF200+MBP+) of brain stem slices with or without Treg cells at 3 d.i.v., Control: MBP+ counts n = 25 FOV and myelination index n = 24 FOV. Treg: MBP+ counts n = 15 and myelination index n = 15 FOV. Fields of view were selected from 3-6 slices/group (MBP+ counts: t = 3.058, d.f. = 38, P = 0.0041; myelination index: t = 3.146, d.f. = 37, P = 0.0033, unpaired, two-tailed, Student’s t test). (d) Representative images of brain slices from (b, c) taken from a z-stack image (scale bar = 100 μm, green = NF200, red = MBP). (e) Analysis of myelination index (NF200+MBP+) of brain stem slices with Treg or Tconv cells at 3 d.i.v. Tconv n = 15 FOV and Treg n = 14 FOV from 3-6 slices per group (U = 26, P = 0.0006, two-tailed Mann-Whitney U test). (f) Representative flow cytometric analysis of non-polarized (NP) T cells and Treg after reactivation for 72h, FSC = Forward Scatter. (g) Analysis of myelination index (NF200+MBP+) of brain stem slices treated with non-polarized (NP) or Treg-conditioned media, at 7 d.i.v. NP n= 14 FOV and Treg n = 15 FOV from 3-6 slices per group (t = 4.559, d.f. = 27, P < 0.0001, unpaired two-tailed Student’s t test). (h-j) Electron microscopic analysis of control (non-demyelinated; M) and remyelinating (RM) brain slices with and without Treg-conditioned medium, n = 3 slices/condition, scale bar = 2 μm (number of myelinated fibres: F(2,6) = 22.89, P = 0.0016; g-ratio: F(2,6) = 5.915, P = 0.0381, one-way ANOVA with Bonferroni post hoc test). Number of myelinated fibres: F = 22.89, Rsquare = 0.8841,C(M) v C(RM) t = 6.592, C(M) v Treg t = 1.978, C(RM) v Treg t = 4.614, P = 0.0016. g-ratio: F = 5.915, R square = 0.6635, C(M) v C(RM) t = 3.344, C(M) v Treg (RM) t = 2.370, p = 0.0381. (k) ELISA of IL-6 in brain slice culture-conditioned media up to day 6 in vitro. n = 4 wells, each from an independent experiment. Data shown are representative of at least 2 (a-e, g) 7 (f) or 1 (h-j) independent experiments. Data presented with mean values indicated, error bars = SEM, *P<0.05, ** P <0.01, *** P <0.001.
Supplementary Figure 3 Treg promote oligodendrocyte differentiation in mixed glia but do not affect OPC survival in the steady state
(a) Immunofluorescence analysis of OPC (green = Olig2, red = Ki67, first row), oligodendrocyte (green = Olig2, red = MBP, second row), astrocyte (green = GFAP, third row) and microglial (red = CD11b, last row) markers after 1 week of mixed glial culture. Blue = DAPI. Left panels depict a stitched image of the well of a 96-well plate (stitched images were taken at 10x) (scale bars from left to right panels = 500 μm, 100 μm, 50 μm). (b, c) Immunofluorescence analysis of MBP+ percentage area (Control vs Treg: U = 4, P = 0.026; Non-Polarized (NP) vs Treg: U = 1, P = 0.0043, two-tailed Mann-Whitney U test) and (c) representative images of mixed glial cultures after 5 days of indicated treatment, n=6, (scale bar = 100 μm, green = Olig2, red = MBP). (d) Immunofluorescence analysis of MBP+ area of mixed glial cultures after 5 days in the presence of Treg-conditioned media prepared from total, or naïve, CD4+ T cells, n = 5 (Control vs Treg: t = 9.786, d.f. = 8, P < 0.0001; control vs naïve Treg: t = 8.644, d.f. = 8, P < 0.0001; Treg vs naïve Treg: t = 1.079, d.f. = 8, P = 0.3119, unpaired two-tailed Student’s t test). (e,f) Immunofluorescence analysis of EthD-1+ cell numbers, n = 5, (U = 10, P = 0.6752, Mann Whitney U test) and (f) representative images of mixed glial cultures (scale bar = 100 μm, green = calcein, red = EthD-1). Data representative of (b) 3 and (d,e) 2 independent experiments. Data presented with mean values indicated, error bars = SEM, * P<0.05, ** P<0.01.
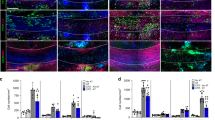
Supplementary Figure 4 Identification of CCN3 as a mediator of Treg-induced oligodendrocyte differentiation
(a) Semi-quantitative proteome profiler analysis of non-polarized (NP) and Treg-conditioned media (2 independent batches) compared to cell culture medium only. (b) Representative immunofluorescence images of mixed glial cultures stained for Olig2+MBP+ cells and analyzed quantitatively in (Figure 4d) (Green = Olig2, red = MBP, scale bar = 100 μm). (c) Schematic diagram of CCN3 depletion and elution from Treg-conditioned media. (d) ELISA confirmation of CCN3 depletion from Treg-conditioned media n = 2. Data representative of 1 (a, with 2 independently generated batches), or at least 3 (b, d) biologically independent experiments.
Supplementary information
Rights and permissions
About this article
Cite this article
Dombrowski, Y., O'Hagan, T., Dittmer, M. et al. Regulatory T cells promote myelin regeneration in the central nervous system. Nat Neurosci 20, 674–680 (2017). https://doi.org/10.1038/nn.4528
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4528
This article is cited by
-
Regulatory T cells limit age-associated retinal inflammation and neurodegeneration
Molecular Neurodegeneration (2024)
-
CD4+CD25+ regulatory T cells ex vivo generated from autologous naïve CD4+ T cells suppress EAE progression
Scientific Reports (2024)
-
Ageing impairs the regenerative capacity of regulatory T cells in mouse central nervous system remyelination
Nature Communications (2024)
-
Transplanted human iPSC-derived vascular endothelial cells promote functional recovery by recruitment of regulatory T cells to ischemic white matter in the brain
Journal of Neuroinflammation (2023)
-
Regulatory T cells alleviate myelin loss and cognitive dysfunction by regulating neuroinflammation and microglial pyroptosis via TLR4/MyD88/NF-κB pathway in LPC-induced demyelination
Journal of Neuroinflammation (2023)