Abstract
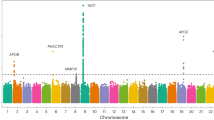
Coronary artery disease (CAD) is a leading cause of morbidity and mortality worldwide1,2. Although 58 genomic regions have been associated with CAD thus far3,4,5,6,7,8,9, most of the heritability is unexplained9, indicating that additional susceptibility loci await identification. An efficient discovery strategy may be larger-scale evaluation of promising associations suggested by genome-wide association studies (GWAS). Hence, we genotyped 56,309 participants using a targeted gene array derived from earlier GWAS results and performed meta-analysis of results with 194,427 participants previously genotyped, totaling 88,192 CAD cases and 162,544 controls. We identified 25 new SNP–CAD associations (P < 5 × 10−8, in fixed-effects meta-analysis) from 15 genomic regions, including SNPs in or near genes involved in cellular adhesion, leukocyte migration and atherosclerosis (PECAM1, rs1867624), coagulation and inflammation (PROCR, rs867186 (p.Ser219Gly)) and vascular smooth muscle cell differentiation (LMOD1, rs2820315). Correlation of these regions with cell-type-specific gene expression and plasma protein levels sheds light on potential disease mechanisms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Roth, G.A. et al. Demographic and epidemiologic drivers of global cardiovascular mortality. N. Engl. J. Med. 372, 1333–1341 (2015).
GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age–sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 385, 117–171 (2015).
CARDIoGRAMplusC4D Consortium. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 45, 25–33 (2013).
Myocardial Infarction Genetics Consortium. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41, 334–341 (2009).
IBC 50K CAD Consortium. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet. 7, e1002260 (2011).
Samani, N.J. et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 357, 443–453 (2007).
Schunkert, H. et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43, 333–338 (2011).
Erdmann, J. et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat. Genet. 41, 280–282 (2009).
CARDIoGRAMplusC4D Consortium. A comprehensive 1000 Genomes–based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47, 1121–1130 (2015).
Voight, B.F. et al. The Metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 8, e1002793 (2012).
Segrè, A.V., Wei, N., Altshuler, D. & Florez, J.C. Pathways targeted by antidiabetes drugs are enriched for multiple genes associated with type 2 diabetes risk. Diabetes 64, 1470–1483 (2015).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Grundberg, E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089 (2012).
Franzén, O. et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science 353, 827–830 (2016).
Ward, L.D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Staley, J.R. et al. PhenoScanner: a database of human genotype–phenotype associations. Bioinformatics 32, 3207–3209 (2016).
Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Teslovich, T.M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
International Consortium for Blood Pressure Genome-Wide Association Studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478, 103–109 (2011).
Surendran, P. et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat. Genet. 48, 1151–1161 (2016).
Zanoni, P. et al. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science 351, 1166–1171 (2016).
Boettger, L.M. et al. Recurring exon deletions in the HP (haptoglobin) gene contribute to lower blood cholesterol levels. Nat. Genet. 48, 359–366 (2016).
Johansson, Å. et al. Identification of genetic variants influencing the human plasma proteome. Proc. Natl. Acad. Sci. USA 110, 4673–4678 (2013).
Holme, I., Aastveit, A.H., Hammar, N., Jungner, I. & Walldius, G. Haptoglobin and risk of myocardial infarction, stroke, and congestive heart failure in 342,125 men and women in the Apolipoprotein MOrtality RISk study (AMORIS). Ann. Med. 41, 522–532 (2009).
Levy, A.P. et al. Haptoglobin genotype is a determinant of iron, lipid peroxidation, and macrophage accumulation in the atherosclerotic plaque. Arterioscler. Thromb. Vasc. Biol. 27, 134–140 (2007).
Westra, H.J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013).
Dennis, J. et al. The endothelial protein C receptor (PROCR) Ser219Gly variant and risk of common thrombotic disorders: a HuGE review and meta-analysis of evidence from observational studies. Blood 119, 2392–2400 (2012).
Tang, W. et al. Genome-wide association study identifies novel loci for plasma levels of protein C: the ARIC study. Blood 116, 5032–5036 (2010).
Smith, N.L. et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation 121, 1382–1392 (2010).
Qu, D., Wang, Y., Song, Y., Esmon, N.L. & Esmon, C.T. The Ser219-->Gly dimorphism of the endothelial protein C receptor contributes to the higher soluble protein levels observed in individuals with the A3 haplotype. J. Thromb. Haemost. 4, 229–235 (2006).
Reiner, A.P. et al. PROC, PROCR and PROS1 polymorphisms, plasma anticoagulant phenotypes, and risk of cardiovascular disease and mortality in older adults: the Cardiovascular Health Study. J. Thromb. Haemost. 6, 1625–1632 (2008).
Uhlen, M. et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 (2010).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Greenawalt, D.M. et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 21, 1008–1016 (2011).
Nanda, V. & Miano, J.M. Leiomodin 1, a new serum response factor–dependent target gene expressed preferentially in differentiated smooth muscle cells. J. Biol. Chem. 287, 2459–2467 (2012).
Chen, J., Kitchen, C.M., Streb, J.W. & Miano, J.M. Myocardin: a component of a molecular switch for smooth muscle differentiation. J. Mol. Cell. Cardiol. 34, 1345–1356 (2002).
Wang, Z., Wang, D.Z., Pipes, G.C. & Olson, E.N. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. USA 100, 7129–7134 (2003).
Kirsten, H. et al. Dissecting the genetics of the human transcriptome identifies novel trait-related trans-eQTLs and corroborates the regulatory relevance of non-protein coding loci. Hum. Mol. Genet. 24, 4746–4763 (2015).
Fairfax, B.P. et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014).
Privratsky, J.R. et al. Relative contribution of PECAM-1 adhesion and signaling to the maintenance of vascular integrity. J. Cell Sci. 124, 1477–1485 (2011).
Harry, B.L. et al. Endothelial cell PECAM-1 promotes atherosclerotic lesions in areas of disturbed flow in ApoE-deficient mice. Arterioscler. Thromb. Vasc. Biol. 28, 2003–2008 (2008).
Goel, R. et al. Site-specific effects of PECAM-1 on atherosclerosis in LDL receptor–deficient mice. Arterioscler. Thromb. Vasc. Biol. 28, 1996–2002 (2008).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013).
Zeller, T. et al. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One 5, e10693 (2010).
Schröder, A. et al. Genomics of ADME gene expression: mapping expression quantitative trait loci relevant for absorption, distribution, metabolism and excretion of drugs in human liver. Pharmacogenomics J. 13, 12–20 (2013).
Schadt, E.E. et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6, e107 (2008).
Lin, H. et al. Gene expression and genetic variation in human atria. Heart Rhythm 11, 266–271 (2014).
Narahara, M. et al. Large-scale East-Asian eQTL mapping reveals novel candidate genes for LD mapping and the genomic landscape of transcriptional effects of sequence variants. PLoS One 9, e100924 (2014).
Innocenti, F. et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 7, e1002078 (2011).
Assimes, T.L. et al. Genetics of coronary artery disease in Taiwan: a cardiometabochip study by the Taichi Consortium. PLoS One 11, e0138014 (2016).
Franceschini, N. et al. Prospective associations of coronary heart disease loci in African Americans using the MetaboChip: the PAGE study. PLoS One 9, e113203 (2014).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Zhou, X. & Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821–824 (2012).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Morris, A.P. Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol. 35, 809–822 (2011).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Zhang, X. et al. Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs. BMC Genomics 15, 532 (2014).
Ernst, J. & Kellis, M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 (2010).
Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012).
Moore, C. et al. The INTERVAL trial to determine whether intervals between blood donations can be safely and acceptably decreased to optimise blood supply: study protocol for a randomised controlled trial. Trials 15, 363 (2014).
Astle, W.J. et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167, 1415–1429 (2016).
Abecasis, G.R. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Arnold, M., Raffler, J., Pfeufer, A., Suhre, K. & Kastenmüller, G. SNiPA: an interactive, genetic variant–centered annotation browser. Bioinformatics 31, 1334–1336 (2015).
Acknowledgements
J.D. is a British Heart Foundation Professor, European Research Council Senior Investigator and NIHR Senior Investigator. J.D.E. and A.D.J. were supported by NHLBI Intramural Research Program funds. N.F. is supported by R21HL123677-01 and R56 DK104806-01A1. N.S. is supported by the British Heart Foundation and is an NIHR Senior Investigator. T.L.A. is supported by NIH career development award K23DK088942. This work was funded by the UK Medical Research Council (G0800270), the British Heart Foundation (SP/09/002), the UK National Institute for Health Research Cambridge Biomedical Research Centre, the European Research Council (268834), European Commission Framework Programme 7 (HEALTH-F2-2012-279233) and Pfizer. The eQTL database construction was supported by NHLBI intramural funds. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute, the National Institutes of Health, or the US Department of Health and Human Services.
A full list of acknowledgments for the studies contributing to this work is provided in the Supplementary Note.
Author information
Authors and Affiliations
Consortia
Contributions
Central analysis group: J.M.M.H., W.Z., D.R.B., T.L.A., A.S.B., D.S. Writing group: J.M.M.H., W.Z., D.R.B., D.S.P., T.L.A., A.S.B., J.D. Study analysts: J.M.M.H., W.-K.H., R.Y., L.L.W., E.L.S., S.F.N., W.-Y.L., R.D., N.F., A.J., A.P.R., C.L.C., K.Y., M.G., D.A., C.A.H., Y.-H.C., X.G., T.L.A. Study PIs and co-PIs: W.H.-H.S., P.D., J.E., S.K., N.J.S., H.S., H.W., D.J.R., J.A.J., S.L.H., A.A.Q., J.S., C.J.P., K.E.N., C.K., U.P., C.A.H., W.-J.L., I.-T.L., R.-H.C., Y.-J.H., J.I.R., J.-M.J.J., T.Q., T.-D.W., D.S.A., A.a.S.M., E.D.A., R.C., Y.-D.I.C., B.G.N., T.L.A., J.D., A.S.B., D.S., A.R., P.F. Bioinformatics, eQTL, pQTL and pathway analyses: D.S.P., W.Z., D.R.B., D.F.F., T.L.A., E.B.F., A.M., J.B.W., E.L.S., B.B.S., A.S.B., J.D.E., A.D.J., P.S., T.L.A., J.M.M.H. Genotyping: S.B., L.A.H., C.K., E.B., U.P., D.A., K.D.T., T.Q., T.L.A. Phenotyping: W.H.H.S., A.T.-H., K.L.R., P.R.K., K.E.N., C.K., C.A.H., W.-J.L., I.-T.L., R.-H.C., Y.-J.H., J.-M.J.J., T.Q., Y.-D.I.C.
Corresponding author
Ethics declarations
Competing interests
A.M., E.B.F. and J.B.W. are full-time employees of Pfizer. D.F.F. is now a full-time employee of Bayer AG, Germany. J.D. reports personal fees and non-financial support from Merck Sharp & Dohme UK Atherosclerosis, the Novartis Cardiovascular & Metabolic Advisory Board, the Pfizer Population Research Advisory Panel and the Sanofi Advisory Board.
Additional information
A full list of members and affiliations appears in the Supplementary Note.
A full list of members and affiliations appears in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 1–4, 6 and 7, and Supplementary Note (PDF 7944 kb)
Supplementary Table 5
Results of meta-analyses across the studies (including all samples) with de novo genotyping at the new CAD-associated SNPs reported in Supplementary Table 4. (XLSX 17 kb)
Supplementary Table 8
Ingenuity Pathway Analysis results. (XLSX 19 kb)
Supplementary Table 9
Coordinates for the genomics regions used for each new CAD locus. (XLSX 9 kb)
Supplementary Table 10
eQTL lookups of the 15 new CAD-associated regions. (XLSX 16 kb)
Supplementary Table 11
HaploReg enrichment analyses of H3K27ac enhancer marks. (XLSX 9 kb)
Supplementary Table 12
Association results for established CAD loci. (XLSX 30 kb)
Rights and permissions
About this article
Cite this article
Howson, J., Zhao, W., Barnes, D. et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet 49, 1113–1119 (2017). https://doi.org/10.1038/ng.3874
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3874
This article is cited by
-
Protein interaction networks in the vasculature prioritize genes and pathways underlying coronary artery disease
Communications Biology (2024)
-
Systemic proteome adaptions to 7-day complete caloric restriction in humans
Nature Metabolism (2024)
-
A comparative analysis of deep learning-based location-adaptive threshold method software against other commercially available software
The International Journal of Cardiovascular Imaging (2024)
-
RBP–RNA interactions in the control of autoimmunity and autoinflammation
Cell Research (2023)
-
Vascular dysfunction caused by loss of Brn-3b/POU4F2 transcription factor in aortic vascular smooth muscle cells is linked to deregulation of calcium signalling pathways
Cell Death & Disease (2023)