Abstract
The grass carp is an important farmed fish, accounting for ∼16% of global freshwater aquaculture, and has a vegetarian diet. Here we report a 0.9-Gb draft genome of a gynogenetic female adult and a 1.07-Gb genome of a wild male adult. Genome annotation identified 27,263 protein-coding gene models in the female genome. A total of 114 scaffolds consisting of 573 Mb are anchored on 24 linkage groups. Divergence between grass carp and zebrafish is estimated to have occurred 49–54 million years ago. We identify a chromosome fusion in grass carp relative to zebrafish and report frequent crossovers between the grass carp X and Y chromosomes. We find that transcriptional activation of the mevalonate pathway and steroid biosynthesis in liver is associated with the grass carp's adaptation from a carnivorous to an herbivorous diet. We believe that the grass carp genome could serve as an initial platform for breeding better-quality fish using a genomic approach.
Similar content being viewed by others
Main
Constituting a member of the Cyprinidae family and the only species of the genus Ctenopharyngodon, the grass carp Ctenopharyngodon idellus is one of the most important aquaculture species, having great commercial value and a worldwide distribution1 (Fig. 1a). Global production of cultured or farmed grass carp is approximately 4.6 million tons per year, accounting for 15.6% of global freshwater aquaculture production in 2011 (ref. 2). The completion of the zebrafish (Danio rerio) genome sequence3 has accelerated studies on the genomes of other members of the Cyprinidae family. In grass carp, progress included the construction of a genetic linkage map4 and the identification of 3,027 UniGene entries5 and 6,269 ESTs6. Such studies have enriched genome research on grass carp. Recent work has focused on genes involved in the immune system7,8, control of food intake9,10, and nutrition and growth11,12. However, the lack of a complete genome sequence has made it difficult to conduct an in-depth investigation of grass carp biology and breeding for better-quality fish. As a first step toward this goal, we report a draft genome sequence and transcriptomic analysis of grass carp, adding this important species to the other sequenced teleosts: cod13, fugu14, medaka15, tetraodon16, stickleback17 and zebrafish3. Taken together, this information provides genomic insights into the evolutionary history of the grass carp and its unique adaptation to a vegetarian diet.
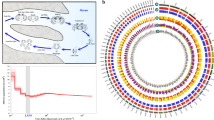
(a) Image of a grass carp adult. (b) Distribution of 55-mer frequency. The distribution of K-mer frequency in the reads was derived from libraries of short insert size (350–400 bp). Values for K-mers are plotted against the frequency (y axis) of their occurrence (x axis). The leftmost truncated peak at low occurrence (1–2) was mainly due to random base errors in the raw sequencing reads. (c) Reconstructed phylogeny of 13 vertebrate genomes. The dN/dS ratio of each branch is shown in blue. The numbers in black correspond to values of bootstrap support. The lamprey is used as an outgroup. Branch length is measured in expected substitutions per site. (d) Venn diagram of gene clusters for five selected vertebrate genomes. Each number represents the number of orthologous gene families shared by the indicated genomes.
Results
Genome assembly
The grass carp genome is composed of 24 pairs of chromosomes18,19 (2n = 48). Adopting a whole-genome shotgun sequencing strategy, we generated approximately 132 Gb of Illumina sequence reads on genomic DNA isolated from the blood of a gynogenetic female adult grass carp and 136 Gb of reads from a wild, water-captured male adult (Supplementary Table 1 and Supplementary Note). We constructed the final assemblies of the female (0.90-Gb) and male (1.07-Gb) genomes using the modified de novo Phusion-meta assembly pipeline, as previously described20 (Supplementary Fig. 1 and Supplementary Table 2). The draft genome of the female was fully annotated to mine genomic information (Fig. 1) and was applied to the anchoring of scaffolds on the genetic linkage map, whereas the male genome was used to detect sequence variation between the male and female genomes.
The female genome assembly had scaffolds with an N50 length greater than 6.4 Mb, and 90% of the assemblies were composed of 301 scaffolds, which were all greater than 179 kb in length (Table 1). Estimation of genome size by distribution of K-mer frequency showed that the female genome was about 891 Mb, close to the size of the assemblies (Fig. 1b and Supplementary Note). We assessed the accuracy of the genome assembly by alignment of the scaffolds to 3,027 published UniGene entires5 and 11 BACs21 (Supplementary Figs. 2 and 3, and Supplementary Table 3), which indicated that the coverage by the initial contigs and scaffolds was approximately 95% and 97%, respectively. Sequence errors were predominantly from insertions or deletions introduced by short-read assembly (Supplementary Table 3a).
We identified a total of 644,817 heterozygous SNPs and 66,101 short indels (10 nucleotides in length or less) in the female genome. In the male genome, we identified 1,465,819 SNPs and 166,867 short indels. The estimated overall heterozygous rates were approximately 0.9 and 2.5 polymorphisms per kilobase in the female and male genomes, respectively (Supplementary Table 4). Clearly, the wild male genome had a much higher heterozygosity rate, which caused a bimodality in the distribution of the K-mer frequency (Fig. 1b) and a shorter length for the assembled scaffolds.
Genome annotation
We annotated a total of 27,263 protein-coding genes in the female genome. The evidence used in gene prediction included 27 Gb of RNA sequencing (RNA-seq) data from 6 tissues (embryo, liver, spleen, brain, kidney and head kidney), over 3,000 known UniGene entries and homologous gene information from zebrafish (Ensembl release 67; Supplementary Fig. 4, Supplementary Table 5 and Supplementary Data Set). We predicted 1,538 tRNA, 24 rRNA, 207 small nucleolar RNA (snoRNA), 136 small nuclear RNA (snRNA) and 444 microRNA genes in our annotation of noncoding RNA genes (Supplementary Tables 6 and 7) and 467,783 simple-sequence repeats (Supplementary Table 8). De novo repeat annotation indicated an overall repeat content of 38%, in comparison to that of 43% in BACs (Supplementary Table 9). This proportion is less than the 52.2% repeat content observed in zebrafish3. This difference might be due to the exclusion of repetitive sequences located in unclosed gaps and on small fragments (<200 bp) of the grass carp assemblies. The majority of transposable elements found in the grass carp genome were type II DNA transposable elements, covering over 20% of the genome, similar to in the zebrafish genome3.
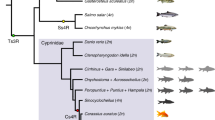
Using the published genetic linkage map of grass carp4, we anchored 114 scaffolds on the 24 linkage groups (Fig. 2a, Supplementary Table 10 and Supplementary Note), covering 573 Mb (64%) of the female assembly with 17,456 (64%) annotated genes localized. Gene synteny over the anchored scaffolds showed that most of the grass carp linkage groups had extensive collinearity with corresponding zebrafish chromosomes (Fig. 2b). Alignment of genes showed high synteny for grass carp and zebrafish, and up to 24,018 grass carp genes (88% of the 27,263 total genes) were located on syntenic blocks (Fig. 2c). Although this result was similar to a previous report4, we found two cross-chromosome arrangements for linkage groups 22 and 24. It is noteworthy that linkage group 24 aligned to zebrafish chromosomes 22 and 10 but not to any other grass carp linkage group. FISH analysis of grass carp chromosomes demonstrated that two grass carp markers aligning to zebrafish chromosomes 10 and 22 were indeed located on the single linkage group 24 (Fig. 2d), explaining why the chromosome number is 25 in zebrafish but 24 in grass carp.
(a) The scaffolds were anchored on a published consensus linkage map4. The blue lines indicate the length of each linkage group (LG) to which the markers are mapped. Map distances between markers are depicted on a Kosambi cM scale. Orange bars represent the anchored scaffolds. The black lines linking markers and scaffolds show the locations of the markers on the scaffolds. The length of each scaffold is shown relative to a 5-Mb scale bar. (b) Syntenic relationship between the zebrafish chromosomes and the grass carp linkage groups. Linkage group 22 is aligned to zebrafish chromosomes 2 and 15, and linkage group 24 is aligned to zebrafish chromosomes 10 and 22. (c) Gene collinearity between zebrafish and grass carp. The zebrafish chromosomes are represented by blue blocks (for example, DR01). The grass carp scaffolds (length > 50 kb) are represented by orange blocks. Aligned genes are connected by green lines. The lengths of the chromosomes and scaffolds are shown relative to a 10-Mb scale bar. (d) FISH study of linkage group 24. The yellow marker CID1435 is located on the region aligned to zebrafish chromosome 10, and the red marker CID0538 is aligned to zebrafish chromosome 22. Scale bar, 5 μm.
Evolutionary analysis
To examine grass carp evolution, we clustered the grass carp gene models with the genes from 12 other vertebrate genomes and used 202 single-copy genes with one-to-one correspondence in the different genomes to reconstruct a phylogenetic tree (Fig. 1c and Supplementary Note). As a species of the Cyprinidae family, grass carp had the closest relationship to zebrafish. According to the TimeTree22 database, the estimated divergence time between zebrafish and grass carp was around 49–54 million years ago (Supplementary Table 11). Most of the selected teleostei genomes showed similar selection pressures, according to calculated dN/dS values (the ratio of the rate of nonsynonymous substitution to the rate of synonymous substitution).
We determined gene families using the TreeFam database23 (Supplementary Note). We performed a five-way comparison among the gene families of a representative mammal (human), bird (chicken), amphibian (frog) and two fishes (zebrafish and grass carp) to quantify the shared or species-specific families present in each genome (Fig. 1d). Zebrafish and grass carp shared 7,227 families, more than the 5,772 families shared by all 5 vertebrate species. Of the 10,184 families identified, 7,171 (70%) carried the same number of gene members in grass carp and zebrafish. Specific comparison of the Hox24,25, Sox26 and Toll-like receptor27 gene clusters among human, medaka, zebrafish and grass carp indicated that the zebrafish and grass carp genomes carried an identical copy number for most subfamilies (Supplementary Figs. 5–7 and Supplementary Table 12). We determined the number of human, zebrafish and grass carp gene members in each family (Supplementary Table 13). The 1,047 families in the class having many grass carp members relative to one human member were composed of 2,658 grass carp genes and 1,047 human genes, with an average ratio of 2.53 grass carp genes to one human gene. Interestingly, the 832 families in the class having many zebrafish members relative to one human member consisted of 2,077 zebrafish genes and 832 human genes, with nearly the same average ratio of 2.50 zebrafish genes to one human gene. It was suggested that the grass carp genome underwent a whole-genome duplication similar to zebrafish after the teleost radiation28.
We estimated the expansion and contraction of gene families to examine their evolutionary history in comparison to the zebrafish, stickleback, tetraodon, fugu, medaka and cod genomes (Supplementary Fig. 8). The significantly expanded families in grass carp included many immune-associated functional domains (P < 0.001; Supplementary Table 14), consistent with the adaption of grass carp to variable environments. Among the 10,184 gene families generated, 2,346 included teleost-specific duplications in zebrafish or grass carp as determined by comparison of the number of gene copies within each family. Of the gene families involved in the teleost-specific duplications, 695 and 295 showed evidence of undergoing a grass carp–specific duplication (GCSD) or a zebrafish-specific duplication (ZSD), respectively (Supplementary Fig. 9 and Supplementary Note), with additional gene duplications found in grass carp and zebrafish. The 695 grass carp families contained 2,561 genes, whereas the 295 zebrafish families consisted of 1,029 genes. We annotated all of these genes using the KEGG29 (Kyoto Encyclopedia of Genes and Genomes) pathway database. Functional analyses of these pathways indicated that genes involved in the ZSD were mainly composed of immune-related genes. Comparably, the grass carp genes involved in the GCSD were not only associated with immune-related genes but also with development-related genes (Fig. 3 and Supplementary Table 15) and were involved in cell proliferation and differentiation (for example, the focal adhesion pathway and the extracellular matrix (ECM)-receptor interaction pathway30,31), nutritional homeostasis (for example, the protein digestion and absorption pathway32,33) and organ size control (for example, the Hippo signaling pathway34,35). Comparison of genes involved in the overview maps of metabolism (reference map ko01100 of the KEGG database) also indicated that grass carp genes involved in the GCSD clustered in carbohydrate metabolism and nucleotide metabolism (Supplementary Fig. 10a), whereas the zebrafish genes only clustered in nucleotide metabolism (Supplementary Fig. 10b). These results indicate that the GCSD was important for adaptation to a vegetarian diet and for some developmental characteristics of grass carp.
All of the genes were involved in the GCSD or ZSD. Pathways were determined by searching the KEGG pathway database. The x axis indicates the numbers of genes involved in each pathway. Pathways with a label in blue highlight metabolic processes that are potentially important to development or diet adaptation.
A potential sex-determination mechanism
By comparison of the assemblies for the male and female grass carp, we identified 206 contigs with a total length of 2.38 Mb that were carried by the male adult but not by the gynogenetic female (Supplementary Fig. 11 and Supplementary Tables 16 and 17). We confirmed each contig by PCR-based sequencing. We also performed PCR amplification of these regions in an extended group of 24 male and 24 female individuals, identifying frequent chromosome crossovers between the X and Y chromosomes in grass carp (Supplementary Fig. 12). Sex in grass carp may be determined not by an entire chromosome but by a few critical genes. Noticeably, we identified a male genome–specific probe that mapped to one of the contigs (probe 184 in Supplementary Fig. 12). We did not find a sequence alignment of this region to any other vertebrate genome, suggesting its unique origin in grass carp.
Gene modeling showed that the male-specific contigs mainly contained genes with domains related to the immunoglobulin V–set, ABC transporter, proteasome subunit and NACHT domains (Supplementary Table 18). Alignment of these predicted genes to the female gene model set showed that 40 genes had homologs in the female genome, of which 22 clustered on linkage group 24 (Supplementary Fig. 13). Gene collinearity and FISH analysis demonstrated that zebrafish chromosomes 10 and 22 fused to form a single chromosome—linkage group 24—in grass carp, which had the largest physical size (Fig. 2d) but the smallest genetic distance, at 23.4 cM, of all of the grass carp linkage groups. Zebrafish has an all-autosome karyotype of 25 chromosomes36, whereas grass carp has differential X and Y chromosomes and a karyotype of 24 chromosomes18,19. Interestingly, linkage group 24 carried most of the predicted genes located in the regions with variable sequence for the male and female genomes, indicating a potential connection between linkage group 24 and a sex-determination chromosome. The identity of the sex-determination chromosome(s) in grass carp is still unclear, but comparison of the male and female genomes may provide further insights into grass carp sex determination, as well as linkage group 24.
Transcriptome analyses of food habit transition
Grass carp are typically herbivorous, a characteristic that has contributed to making them a popular breeding species. How grass carp effectively absorb nutrients from plants to support their rapid growth is an unanswered research question37. Grass carp complete the transition from a carnivorous to an herbivorous diet when they become 3 to 5.5 cm in body length, around 1.5 months after hatching. We analyzed RNA-seq data derived from gut, liver and brain to characterize variations in gene expression before and after the change (Fig. 4a and Online Methods). Genes with differential expression were significantly enriched in pathways associated with circadian rhythm in gut and with steroid biosynthesis, terpenoid backbone biosynthesis and glycerophospholipid metabolism pathways in liver (DAVID Bioinformatics Resources38; P < 0.05; Supplementary Table 19).
(a) Design of the FHT experiments. At 46 d after hatching, fish not undergoing FHT (fed with chironomid larvae) were collected as sample “46 d”. During the period from 46 to 116 d after hatching, the fish were divided into two groups—one fed with duckweed and the other still fed with chironomid larvae—which were collected at 116 d after hatching as sample “116 d–FHT” and sample “116 d,” respectively. (b) Activation of the mevalonate pathway and steroid biosynthesis. The blue and orange arrows indicate the reaction steps in different pathways. The number in each rectangle shows the EC code of the enzyme catalyzing that transfer. Red codes indicate enzyme genes with significantly increased expression after the FHT (q < 0.001). Gene expression as measured by quantification of transcription levels (reads per kilobase of exon model per million mapped reads, RPKM45) is shown in the histograms. Compound names are shown beside the corresponding circles.
After the change in diet, it is essential for grass carp to maintain a continuous feeding rhythm so that they can obtain sufficient nutrients from their food. The analyses of liver RNA-seq data identified significant activation of the steroid biosynthesis pathway downstream of the mevalonate pathway in terpenoid backbone biosynthesis, as reflected by an average expression level 32-fold higher than before the change in diet (q value39 < 0.001; Fig. 4b). In comparison, control individuals not undergoing a change in diet had an average 0.94-fold increase in the expression levels of these genes (Supplementary Table 19b,c). We confirmed the differential expression levels of most of these genes by quantitative RT-PCR (qRT-PCR; Supplementary Fig. 14), finding an average 58-fold increase in expression after the change in diet. The mevalonate pathway is employed to transfer the products of glycolysis to precursors of terpenoids (such as farnesyl-PP), which can potentially be metabolized by downstream steroid biosynthesis pathways and used for different processes, such as the biosynthesis of hormones and vitamin D40,41. During the diet change experiment, we observed that grass carp fed with duckweed had considerably greater growth than those fed with chironomid larvae (fish before the transition and fish not undergoing a transition) in terms of body length, gut length, body weight and the rate of gut length/body length (data not shown). Metabolic adaption of these pathways apparently supports the effective use of plant-derived nutrients in grass carp.
In addition, a previous report indicated that grass carp fed with a plant diet spend a longer time on feeding37. We also observed that the grass carp that received a plant diet fed almost continuously throughout any given 24-h period. Analyses of transcriptome data showed that genes involved in the circadian rhythm pathway were activated after the food habit transition (FHT), which included the clock-bmal1 heterodimer and the ror and clock genes (Supplementary Fig. 15a). The related genes showed high similarity to those of zebrafish (such as the rorca42 and clocka43 genes), with the exception of some promoter regions of genes carrying different elements (Supplementary Fig. 15b). Although the relationship between the genes involved in circadian rhythm and continuous feeding is unclear, this finding might suggest that feeding frequency should be examined during the diet change of grass carp.
We investigated the non–grass carp reads for potential gut microbiota by removing all gut RNA-seq reads with alignment to the grass carp assemblies (Supplementary Tables 20 and 21, and Supplementary Note). This analysis did not identify any genes encoding predicted cellulose-digesting enzymes in the gut, suggesting that the grass carp intestine may not digest and absorb cellulose, which reinforces the view that cellulase may be developed as an aquatic additive to promote the growth of grass carp44. However, sequencing of gut biota will likely prove to be more powerful in understanding how the grass carp digests plant materials.
Discussion
We have generated two draft grass carp genome assemblies, a male one and a fully annotated female assembly. Comparison of a large number of gene models and synteny analysis demonstrated that zebrafish and grass carp share a similar genomic evolutionary history. However, a chromosome fusion resulting in linkage group 24 and the occurrence of a GCSD may be responsible for the substantial differences in development (for example, in body size) and other characteristics (for example, sex determination) between grass carp and zebrafish. Characterization of diet change–associated transcriptomes has provided new genomic information on the metabolic adaption of grass carp to the shift to a vegetarian diet during its life history. These genome sequences also bring grass carp breeding into a genomic phase.
Methods
DNA library preparation and sequencing.
Genomic DNA was isolated from the blood cells of a gynogenetic female adult grass carp and a wild-captured male adult grass carp using the DNeasy Blood and Tissue kit (Qiagen). An amplification-free approach46 was applied to prepare sequencing libraries with short inserts of 350–450 bp for paired-end reads, following the manufacturer's protocol for Illumina. To construct the libraries with insert sizes of 1, 3, 5 and 8–10 kb for mate-paired reads (Supplementary Fig. 16), the protocols of the Mate-Pair Library v2 Sample Preparation Guide (Illumina) and the Paired-End Library Preparation Method Manual (Roche) were combined. Raw data were generated by an Illumina HiSeq 2000 sequencer and an Illumina Genome Analyzer IIx sequencer.
Sequence assembly.
A de novo assembly pipeline was developed to assemble the short reads (Supplementary Fig. 1), which was a modified version of the Phusion-meta method as previously reported20. Briefly, when the paired-end reads were screened to remove low-quality reads containing ten or more unique K-mers, they were clustered into thousands of groups by Phusion2 with K-mer set at 51 bp. The reads grouped into each cluster were assembled in parallel into contigs by ABYSS47, fermi48 and SOAPdenovo49. All these contigs were merged to form initial draft contigs. Consensus contigs were obtained by aligning all the reads back to the draft contigs using the assembly management tool GAP5 (ref. 50). The mate-paired reads were then hierarchically and iteratively assembled into contigs to build preliminary scaffolds with SOAPdenovo (K-mer set at 61 bp). Final scaffolding was conducted with Spinner.
Prediction of protein-coding genes.
The construction of gene models was based on expression sequences (UniGene entries and RNA-seq data) and homologs of zebrafish genes (Ensembl release 67). We built a seven-step pipeline to facilitate the gene model set. (1) A total of 3,027 published UniGene sequences were mapped to the repeat-masked scaffolds by an mRNA/EST genome mapping program in GMAP51 with the parameters set to ‘-n 1 -f 2 -B 1 -A -t 4’. (2) Protein sequences form zebrafish were aligned to the grass carp genome using TBLASTN with an E-value cutoff of 1 × 10−10. (3) The Illumina RNA-seq sequences from six organs or tissues (embryo, liver, spleen, brain, kidney and head kidney) were mapped to the repeat-masked scaffolds using the SMALT aligner with the parameters set to a minimum Smith-Waterman (-m) value of 35, a maximum insert size (-i) of 5,000 and a minimum insert size (-j) of 20. Only uniquely matched reads were selected to assist in gene prediction. (4) On the basis of the alignment results from steps 1–3, the target gene fragments found by extending the aligned regions by 3,000 bp on both ends were selected to build preliminary gene models with FgeneSH++ (Softberry) using parameters trained on fish genomes. If an overlap was found for adjacent regions, the fragments were selected in order of priority for alignment from UniGene sequences to zebrafish homologs to transcriptome sequences. (5) To check the accuracy of the preliminary gene models, the models were aligned to the zebrafish gene model set with BLASTP with an E-value cutoff of 1 × 10−20. When a grass carp gene mapped to two or more adjacent zebrafish genes, this chimeric gene was split according to information from the zebrafish homologs. When two or more adjacent grass carp genes were aligned to a single zebrafish gene, they were combined by Genewise52 using the corresponding zebrafish gene as the reference sequence. The longest translation product was chosen to represent each gene. (6) The coding sequence for each gene model was aligned to the Repbase library and a grass carp repeat library created with RepeatModeler, using BLASTN results with an E-value cutoff of 1 × 10−5, to remove transposon genes. (7) Genes with a coding sequence of less than 60 amino acids were removed. The genes comprising three or fewer exons and not supported by expression sequences or zebrafish homologs were also removed. In this manner, the grass carp gene models were generated.
Transcriptome sequencing.
Six tissues (embryo, liver, spleen, brain, kidney and head kidney) were collected from a male adult to perform RNA-seq, with the resulting data used for gene modeling. All these samples, as well as those from the FHT experiments, were subjected to RNA isolation using SV TRIzol Reagent (Invitrogen). Poly(A)+ mRNA was purified using the DynaBeads mRNA Purification kit (Life Technologies). Paired-end cDNA libraries were constructed using the RNA Seq NGS Library Preparation Kit for Whole-Transcriptome Discovery (Gnomegen). The resulting paired-end cDNA libraries were sequenced using the Illumina HiSeq 2000 system.
Identification of male contigs located on the potential sex-determination chromosome.
Male contigs (length ≥ 500 bp) were aligned to the female contigs (length ≥ 500 bp) using the MUMmer53 aligner with default parameters. Coverage of each male contig by the female ones was then estimated using an identity threshold of ≥95% and requiring that any unaligned gap within the aligned region be ≤10 bp in length. Male contigs with coverage less than 0.05 were selected to be masked by RepeatMasker against the grass carp repetitive sequence library. The masked contigs were used to design the primers for the following PCR analysis. Using male and female genomic DNA as template, the primer pairs primed PCR amplification by Easy Taq DNA Polymerase (Transgen Biotech) to verify each of the contigs. Primer sequences are listed in Supplementary Table 16. Male contigs with amplification from male DNA but no amplification from female DNA were confirmed by sequencing of the PCR products.In total, 206 contigs were identified that potentially carried sequence variation between the male and female genomes, and these contigs were annotated to predict protein-coding genes using FgeneSH++.
Sample preparation and identification of differentially expressed genes in the FHT experiments.
Grass carp larvae were raised in tanks at 25 ± 2 °C and fed with chironomid larvae (Chironomus tentans). At 46 d after hatching (body weight = 0.39 ± 0.05 g, body length = 28.05 ± 0.99 mm), 30 fish were randomly selected for sample collection before the FHT (designated as “46 d”). The rest of the fish were then randomly divided into 2 groups (n = 1,000 for each group) and fed with either chironomid larvae as fish undergoing no transition (designated as “116 d”) or duckweed Lemna minor (designated as “116 d–FHT”) as fish undergoing a transition to herbivory. Fish had free access to food 24 h a day and fed for 70 d. At 116 d after hatching (body weight = 2.97−7.34 g, body length = 53.96−72.78 mm), 30 fish were randomly selected from each group for sample collection. The collection time for all samples was at 9 a.m. Total RNA was isolated from brain, liver and gut tissues using SV TRIzol Reagent (Invitrogen) according to the manufacturer's protocol. Equal amounts of total RNA from nine fish in each group were pooled for each tissue and used to construct the libraries for transcriptome analysis.
The resulting RNA-seq reads were aligned to the gene model set with the SMALT aligner using the parameters of a minimum Smith-Waterman (-m) value of 35, a maximum insert size (-i) of 5,000 and a minimum insert size (-j) of 20. Numbers of uniquely mapped reads (mapping score ≥ 50) were converted to quantified transcript levels by RPKM. The R package DEGseq54 was used to digitally measure differential expression at the annotated loci. When a gene's expression increased or decreased in 116 d–FHT liver by more than twofold (q value39 < 0.001) compared to that in the 46 d sample (before FHT), Δ1 was defined as log2 (RPKM(116 d–FHT)/RPKM (46 d)). When a gene's expression increased or decreased in 116 d liver (not FHT) by more than twofold compared to that in the 46 d sample (q value < 0.001), Δ2 was defined as log2 (RPKM(116 d)/RPKM(46 d)). If the value of Δ1 – Δ2 was less than −1 or greater than 1, the corresponding gene was considered to be differentially expressed by the FHT and non-FHT samples. All identified genes were functionally annotated by DAVID to group them into significantly clustered pathways (P < 0.05). Ethical approval for this work was obtained from the Expert Committee of Biomedical Ethics, Institute of Hydrobiology of the Chinese Academy of Sciences (Y11201-1-301).
URLs.
Kyoto Encyclopedia of Genes and Genomes (KEGG), http://www.genome.jp/kegg/; KEGG Automatic Annotation Server (KAAS), http://www.genome.jp/tools/kaas/; DAVID, http://david.abcc.ncifcrf.gov/summary.jsp; SMALT, http://www.sanger.ac.uk/resources/software/smalt/; SOAPdenovo, http://soap.genomics.org.cn/; RepeatModeler, http://www.repeatmasker.org/RepeatModeler.html; RepeatMasker, http://www.repeatmasker.org/; Repbase, http://www.girinst.org/repbase/; TimeTree, http://www.timetree.org/; TreeFam, http://treefam.genomics.org.cn/; fermi, https://github.com/lh3/fermi; Spinner, ftp://ftp.sanger.ac.uk/pub/users/zn1/spinner/; Ensembl, ftp://ftp.ensembl.org/pub/; PHYLIP, http://evolution.genetics.washington.edu/phylip.html.
Accession codes.
All of the Illumina short-read sequencing data for this project have been deposited at the DNA Data Bank of Japan (DDBJ), the European Molecular Biology Laboratory (EMBL) and GenBank under accession PRJEB5920, which includes whole-genome shotgun sequence (ERS428355 for the female and ERS428356 for the male; Sequence Read Archive (SRA)) and RNA-seq data (ERS430059, SRA). All of the grass carp data have been released at the official National Center for Gene Research website (http://www.ncgr.ac.cn/grasscarp/). The current version of the data set is the first version (CI01).
Change history
15 July 2015
In the version of this article initially published, the affiliation for Songhun Jang was incorrectly listed as Kim Illinois Sung University, Pyongyang, North Korea. The correct affiliation is Kim Il Sung University, Pyongyang, North Korea. The error has been corrected in the HTML and PDF versions of the article.
References
Chilton, E.W. & Muoneke, M.I. Biology and management of grass carp (Ctenopharyngodon idella, Cyprinidae) for vegetation control: a North American perspective. Rev. Fish Biol. Fish. 2, 283–320 (1992).
Statistics and Information Service, Fisheries and Aquaculture Department, Food and Agriculture Organization of the United Nations. FAO Yearbook of Fishery and Aquaculture Statistics 2011 (Food and Agriculture Organization of the United Nations, 2013).
Howe, K. et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 (2013).
Xia, J.H. et al. A consensus linkage map of the grass carp (Ctenopharyngodon idella) based on microsatellites and SNPs. BMC Genomics 11, 135 (2010).
Chen, J. et al. Transcriptome analysis of head kidney in grass carp and discovery of immune-related genes. BMC Vet. Res. 8, 108 (2012).
Xu, B. et al. Generation and analysis of ESTs from the grass carp, Ctenopharyngodon idellus. Anim. Biotechnol. 21, 217–225 (2010).
Guo, T. et al. Cloning, molecular characterization, and expression analysis of the signal transducer and activator of transcription 3 (STAT 3) gene from grass carp (Ctenopharyngodon idellus). Fish Shellfish Immunol. 35, 1624–1634 (2013).
Wang, T.T. et al. Molecular characterization, expression analysis, and biological effects of interleukin-8 in grass carp Ctenopharyngodon idellus. Fish Shellfish Immunol. 35, 1421–1432 (2013).
Chen, Y., Pandit, N.P., Fu, J., Li, D. & Li, J. Identification, characterization and feeding response of peptide YYb (PYYb) gene in grass carp (Ctenopharyngodon idellus). Fish Physiol. Biochem. 40, 45–55 (2014).
Feng, K., Zhang, G.R., Wei, K.J. & Xiong, B.X. Molecular cloning, tissue distribution, and ontogenetic expression of ghrelin and regulation of expression by fasting and refeeding in the grass carp (Ctenopharyngodon idellus). J. Exp. Zool. A Ecol. Genet. Physiol. 319, 202–212 (2013).
Yu, E.M. et al. Molecular cloning of type I collagen cDNA and nutritional regulation of type I collagen mRNA expression in grass carp. J. Anim. Physiol. Anim. Nutr. (Berl.) 98, 755–765 (2014).
Zhong, S.S., Jiang, X.Y., Sun, C.F. & Zou, S.M. Identification of a second follistatin gene in grass carp (Ctenopharyngodon idellus) and its regulatory function in myogenesis during embryogenesis. Gen. Comp. Endocrinol. 185, 19–27 (2013).
Star, B. et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature 477, 207–210 (2011).
Aparicio, S. et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297, 1301–1310 (2002).
Kasahara, M. et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 447, 714–719 (2007).
Jaillon, O. et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946–957 (2004).
Jones, F.C. et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61 (2012).
Nocusa, S. A comparative study of the chromosomes in the fishes with particular consideration on taxonomy and evolution. Mem. Hyogo Univ. Agric. 3, 1–62 (1960).
Stanley, J.G. Female homogamety in grass carp (Ctenopharyngodon idella) determined by gynogenesis. J. Fish. Res. Board Can. 33, 1372–1374 (1976).
Peng, Z. et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 45, 456–461 (2013).
Jang, S. et al. Construction and characterization of two bacterial artificial chromosome libraries of grass carp. Mar. Biotechnol. (NY) 12, 261–266 (2010).
Kumar, S. & Hedges, S.B. TimeTree2: species divergence times on the iPhone. Bioinformatics 27, 2023–2024 (2011).
Li, H. et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 34, D572–D580 (2006).
Crow, K.D., Stadler, P.F., Lynch, V.J., Amemiya, C. & Wagner, G.P. The “fish-specific” Hox cluster duplication is coincident with the origin of teleosts. Mol. Biol. Evol. 23, 121–136 (2006).
Kurosawa, G. et al. Organization and structure of hox gene loci in medaka genome and comparison with those of pufferfish and zebrafish genomes. Gene 370, 75–82 (2006).
Wegner, M. All purpose Sox: the many roles of Sox proteins in gene expression. Int. J. Biochem. Cell Biol. 42, 381–390 (2010).
Akira, S., Yamamoto, M. & Takeda, K. Role of adapters in Toll-like receptor signalling. Biochem. Soc. Trans. 31, 637–642 (2003).
Braasch, I. & Postlethwait, J.H. in. Polyploidy and Genome Evolution (eds. Soltis, P.S. and Soltis, D.E.) 341–383 (Springer, 2012).
Kanehisa, M. et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 (2014).
Petit, V. & Thiery, J.P. Focal adhesions: structure and dynamics. Biol. Cell 92, 477–494 (2000).
Danen, E.H. & Yamada, K.M. Fibronectin, integrins, and growth control. J. Cell. Physiol. 189, 1–13 (2001).
Palacín, M. et al. The genetics of heteromeric amino acid transporters. Physiology (Bethesda) 20, 112–124 (2005).
Goodman, B.E. Insights into digestion and absorption of major nutrients in humans. Adv. Physiol. Educ. 34, 44–53 (2010).
Zhao, B., Tumaneng, K. & Guan, K.L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 13, 877–883 (2011).
Zhao, B., Lei, Q.Y. & Guan, K.L. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 20, 638–646 (2008).
Traut, W. & Winking, H. Meiotic chromosomes and stages of sex chromosome evolution in fish: zebrafish, platyfish and guppy. Chromosome Res. 9, 659–672 (2001).
Cui, Y., Chen, S., Wang, S. & Liu, X. Laboratory observations on the circadian feeding patterns in the grass carp (Ctenopharyngodon idella Val.) fed three different diets. Aquaculture 113, 57–64 (1993).
Huang, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 4, 44–57 (2009).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57, 289–300 (1995).
Dempsey, M.E. Regulation of steroid biosynthesis. Annu. Rev. Biochem. 43, 967–990 (1974).
Lange, B.M., Rujan, T., Martin, W. & Croteau, R. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. USA 97, 13172–13177 (2000).
Flores, M.V. et al. The zebrafish retinoid-related orphan receptor (ror) gene family. Gene Expr. Patterns 7, 535–543 (2007).
Tan, Y., DeBruyne, J., Cahill, G.M. & Wells, D.E. Identification of a mutation in the Clock1 gene affecting zebrafish circadian rhythms. J. Neurogenet. 22, 149–166 (2008).
Zhou, Y. et al. Enhancement of growth and intestinal flora in grass carp: the effect of exogenous cellulase. Aquaculture 416–417, 1–7 (2013).
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628 (2008).
Kozarewa, I. et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods 6, 291–295 (2009).
Simpson, J.T. et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123 (2009).
Li, H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics 28, 1838–1844 (2012).
Li, R. et al. SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714 (2008).
Bonfield, J.K. & Whitwham, A. Gap5—editing the billion fragment sequence assembly. Bioinformatics 26, 1699–1703 (2010).
Wu, T.D. & Watanabe, C.K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875 (2005).
Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Res. 14, 988–995 (2004).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Wang, L., Feng, Z., Wang, X., Wang, X. & Zhang, X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138 (2010).
Acknowledgements
We thank S. Liu (Hunan Normal University) for providing a gynogenetic female individual. We thank Y. Xue and J. Torrance for their help in English-language editing of the manuscript. This work is supported by the National High-Technology Research and Development Program (863 Program, 2011AA100403), the National Natural Science Foundation of China (31130055), the Strategic Pilot Science and Technology Projects (A) Category, Chinese Academy of Science (XDA08030203), the Guangdong Provincial Science and Technology Program (2012B090500008) and the State Key Laboratory of Freshwater Ecology and Biotechnology (2011FBZ18).
Author information
Authors and Affiliations
Contributions
Z.Z., B.H. and H. Lin conceived and managed the project and its components and contributed to the original concepts of the project. Ying Lu and Yan Li designed and performed genome annotation and RNA-seq data analysis. Zemin Ning, H. Lu and Q.Z. developed the de novo assembly pipeline and performed de novo genome assembly. Y.W. and Y.Z. designed the biological experiments. Sample preparation and genome sequencing were carried out by Q.F., D.F., Q.W., Yiqi Lu, W.L., Z.W., C. Zhou, Q.T., R.H., F.D., W.H., L.X., L. Liao and Yongming Li. T.L., L.Z., K.L., C. Zhu, S.L., L.H., C.Y., X.K., X.X., W.Z. and M.S. participated in analyses of RNA-seq data and in gene family, SNP and functional annotation. T.H. provided IT support. X. Liang and S.H. designed and performed the FHT experiment. Ying Lu, Q.F., Yiqi Lu, Z.W., H.H., X. Liu and Zhen Ning identified the contigs carrying sequence variation between the male and female individuals. B.G. and S.J. carried out BAC library construction. R.Y., Q.L. and L. Luo conducted the FISH analysis. Ying Lu and B.H. analyzed the data as a whole and wrote the manuscript. Z.Z., H. Lin and Y.W. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Tables 1–15 and 19–21, and Supplementary Figures 1–16. (PDF 8276 kb)
Supplementary Table 16
Primers used in identification of the presence or absence of the sequences differing between the sequenced gynogenetic female and male adults. (XLSX 21 kb)
Supplementary Table 17
List of 206 male contigs and their location on the male scaffolds. (XLSX 106 kb)
Supplementary Table 18
Predicted genes and their functional annotation in the male contigs carrying potential sequencing differences between the male and female. (XLSX 17 kb)
Supplementary Data Set
Supplementary Data Set (XLSX 1966 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/.
About this article
Cite this article
Wang, Y., Lu, Y., Zhang, Y. et al. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat Genet 47, 625–631 (2015). https://doi.org/10.1038/ng.3280
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3280
This article is cited by
-
Deciphering the gut microbiome of grass carp through multi-omics approach
Microbiome (2024)
-
Ran in Procambarus clarkii: molecular characterization and immune function
Aquaculture International (2024)
-
The chromosome-level genome and key genes associated with mud-dwelling behavior and adaptations of hypoxia and noxious environments in loach (Misgurnus anguillicaudatus)
BMC Biology (2023)
-
Transcriptome Analysis of the Spleen Provides Insight into the Immune Regulation of GCRV Resistance in Grass Carp (Ctenopharyngodon idella)
Marine Biotechnology (2023)
-
Variations in the Behaviour, Survival, Haematology, and Biochemistry of Ctenopharyngodon idella (Grass Carp) After Exposure to Commercial Grade Atrazine
Bulletin of Environmental Contamination and Toxicology (2023)