Abstract
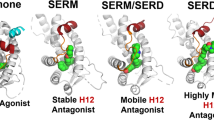
Resistance to endocrine therapies remains a major clinical problem for the treatment of estrogen receptor-α (ERα)-positive breast cancer. On-target side effects limit therapeutic compliance and use for chemoprevention, highlighting an unmet need for new therapies. Here we present a full-antagonist ligand series lacking the prototypical ligand side chain that has been universally used to engender antagonism of ERα through poorly understood structural mechanisms. A series of crystal structures and phenotypic assays reveal a structure-based design strategy with separate design elements for antagonism and degradation of the receptor, and access to a structurally distinct space for further improvements in ligand design. Understanding structural rules that guide ligands to produce diverse ERα-mediated phenotypes has broad implications for the treatment of breast cancer and other estrogen-sensitive aspects of human health including bone homeostasis, energy metabolism, and autoimmunity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
17 March 2017
In the version of this article initially published, Figure 1 was missing the ring designation letters, the substituent designation (R1/R2) and the ring locant numbers for the compounds in panel a. The error has been corrected in the HTML and PDF versions of the article.
17 March 2017
In the version of this article initially published, some of the chemical compounds were incorrectly numbered as stereoisomers instead of racemates and some of the stereoisomers were marked with incorrect stereocenter configurations. In numerous instances throughout the main text and Online Methods, compounds 1–13 were incorrectly called 1R–13R (6 instances of 13R are correct and were left unchanged). In the legend for Figure 5: 1S, 4S, 11S, 12S, 13S were incorrectly called 1R, 4R, 11R, 12R, 13R, respectively; in the left column of p.115: 1S, 4S, 11S were incorrectly called 1R, 4R, 11R, respectively; and in the left column of p.116: 13S was incorrectly called 13R. The errors have been corrected in the HTML and PDF versions of the article.
References
Yu, K.D., Wu, J., Shen, Z.Z. & Shao, Z.M. Hazard of breast cancer-specific mortality among women with estrogen receptor-positive breast cancer after five years from diagnosis: implication for extended endocrine therapy. J. Clin. Endocrinol. Metab. 97, E2201–E2209 (2012).
Dunnwald, L.K., Rossing, M.A. & Li, C.I. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 9, R6 (2007).
Kennecke, H. et al. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 28, 3271–3277 (2010).
Lai, A. et al. Identification of GDC-0810 (ARN-810), an orally bioavailable selective estrogen receptor degrader (SERD) that demonstrates robust activity in tamoxifen-resistant breast cancer xenografts. J. Med. Chem. 58, 4888–4904 (2015).
Wardell, S.E., Nelson, E.R., Chao, C.A., Alley, H.M. & McDonnell, D.P. Evaluation of the pharmacological activities of RAD1901, a selective estrogen receptor degrader. Endocr. Relat. Cancer 22, 713–724 (2015).
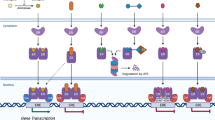
McKenna, N.J., Lanz, R.B. & O'Malley, B.W. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20, 321–344 (1999).
Métivier, R. et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763 (2003).
Cicatiello, L. et al. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol. Cell. Biol. 24, 7260–7274 (2004).
Brzozowski, A.M. et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389, 753–758 (1997).
Shiau, A.K. et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95, 927–937 (1998).
Pike, A.C. et al. Structural insights into the mode of action of a pure antiestrogen. Structure 9, 145–153 (2001).
Wu, Y.L. et al. Structural basis for an unexpected mode of SERM-mediated ER antagonism. Mol. Cell 18, 413–424 (2005).
Willson, T.M. et al. Dissection of the molecular mechanism of action of GW5638, a novel estrogen receptor ligand, provides insights into the role of estrogen receptor in bone. Endocrinology 138, 3901–3911 (1997).
Laxmi, Y.R. et al. Anti-breast cancer potential of SS1020, a novel antiestrogen lacking estrogenic and genotoxic actions. Int. J. Cancer 127, 1718–1726 (2010).
Shiau, A.K. et al. Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism. Nat. Struct. Biol. 9, 359–364 (2002).
Nettles, K.W. et al. NF kappa B selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses. Nat. Chem. Biol. 4, 241–247 (2008).
Zheng, Y. et al. Development of selective estrogen receptor modulator (SERM)-like activity through an indirect mechanism of estrogen receptor antagonism: defining the binding mode of 7-oxabicyclo[2.2.1]hept-5-ene scaffold core ligands. ChemMedChem 7, 1094–1100 (2012).
Zhu, M. et al. Bicyclic core estrogens as full antagonists: synthesis, biological evaluation and structure-activity relationships of estrogen receptor ligands based on bridged oxabicyclic core arylsulfonamides. Org. Biomol. Chem. 10, 8692–8700 (2012).
Kastrati, I., Canestrari, E. & Frasor, J. PHLDA1 expression is controlled by an estrogen receptor-NFκB-miR-181 regulatory loop and is essential for formation of ER+ mammospheres. Oncogene 34, 2309–2316 (2015).
Cvoro, A. et al. Distinct roles of unliganded and liganded estrogen receptors in transcriptional repression. Mol. Cell 21, 555–564 (2006).
Nettles, K.W. et al. CBP Is a dosage-dependent regulator of nuclear factor-kappaB suppression by the estrogen receptor. Mol. Endocrinol. 22, 263–272 (2008).
Chinenov, Y., Gupte, R. & Rogatsky, I. Nuclear receptors in inflammation control: repression by GR and beyond. Mol. Cell. Endocrinol. 380, 55–64 (2013).
Nwachukwu, J.C. et al. Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network. eLife 3, e02057 (2014).
Srinivasan, S. et al. Ligand-binding dynamics rewire cellular signaling via estrogen receptor-α. Nat. Chem. Biol. 9, 326–332 (2013).
Nwachukwu, J.C. et al. Predictive features of ligand-specific signaling through the estrogen receptor. Mol. Syst. Biol. 12, 864 (2016).
Carlson, K.E., Choi, I., Gee, A., Katzenellenbogen, B.S. & Katzenellenbogen, J.A. Altered ligand binding properties and enhanced stability of a constitutively active estrogen receptor: evidence that an open pocket conformation is required for ligand interaction. Biochemistry 36, 14897–14905 (1997).
Nettles, K.W. et al. Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO Rep. 8, 563–568 (2007).
Delfosse, V. et al. Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes. Proc. Natl. Acad. Sci. USA 109, 14930–14935 (2012).
Bruning, J.B. et al. Coupling of receptor conformation and ligand orientation determine graded activity. Nat. Chem. Biol. 6, 837–843 (2010).
Jeselsohn, R. et al. Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin. Cancer Res. 20, 1757–1767 (2014).
Li, S. et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 4, 1116–1130 (2013).
Robinson, D.R. et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 45, 1446–1451 (2013).
Toy, W. et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 45, 1439–1445 (2013).
Patani, N. & Martin, L.A. Understanding response and resistance to oestrogen deprivation in ER-positive breast cancer. Mol. Cell. Endocrinol. 382, 683–694 (2014).
Kuske, B. et al. Endocrine therapy resistance can be associated with high estrogen receptor alpha (ERalpha) expression and reduced ERalpha phosphorylation in breast cancer models. Endocr. Relat. Cancer 13, 1121–1133 (2006).
Billon-Galés, A. et al. The transactivating function 1 of estrogen receptor alpha is dispensable for the vasculoprotective actions of 17beta-estradiol. Proc. Natl. Acad. Sci. USA 106, 2053–2058 (2009).
Abot, A. et al. The AF-1 activation function of estrogen receptor α is necessary and sufficient for uterine epithelial cell proliferation in vivo. Endocrinology 154, 2222–2233 (2013).
Bhatt, S., Xiao, Z., Meng, Z. & Katzenellenbogen, B.S. Phosphorylation by p38 mitogen-activated protein kinase promotes estrogen receptor α turnover and functional activity via the SCF(Skp2) proteasomal complex. Mol. Cell. Biol. 32, 1928–1943 (2012).
Fan, M., Bigsby, R.M. & Nephew, K.P. The NEDD8 pathway is required for proteasome-mediated degradation of human estrogen receptor (ER)-alpha and essential for the antiproliferative activity of ICI 182,780 in ERalpha-positive breast cancer cells. Mol. Endocrinol. 17, 356–365 (2003).
Lupien, M. et al. Raloxifene and ICI182,780 increase estrogen receptor-alpha association with a nuclear compartment via overlapping sets of hydrophobic amino acids in activation function 2 helix 12. Mol. Endocrinol. 21, 797–816 (2007).
Andruska, N.D. et al. Estrogen receptor α inhibitor activates the unfolded protein response, blocks protein synthesis, and induces tumor regression. Proc. Natl. Acad. Sci. USA 112, 4737–4742 (2015).
Connor, C.E. et al. Circumventing tamoxifen resistance in breast cancers using antiestrogens that induce unique conformational changes in the estrogen receptor. Cancer Res. 61, 2917–2922 (2001).
Wardell, S.E., Marks, J.R. & McDonnell, D.P. The turnover of estrogen receptor α by the selective estrogen receptor degrader (SERD) fulvestrant is a saturable process that is not required for antagonist efficacy. Biochem. Pharmacol. 82, 122–130 (2011).
Wardell, S.E., Nelson, E.R., Chao, C.A. & McDonnell, D.P. Bazedoxifene exhibits antiestrogenic activity in animal models of tamoxifen-resistant breast cancer: implications for treatment of advanced disease. Clin. Cancer Res. 19, 2420–2431 (2013).
Wittmann, B.M., Sherk, A. & McDonnell, D.P. Definition of functionally important mechanistic differences among selective estrogen receptor down-regulators. Cancer Res. 67, 9549–9560 (2007).
Dontu, G. et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 (2003).
Gupta, P.B. et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138, 645–659 (2009).
Frasor, J. et al. Response-specific and ligand dose-dependent modulation of estrogen receptor (ER) alpha activity by ERbeta in the uterus. Endocrinology 144, 3159–3166 (2003).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006).
Adams, P.D. et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011).
Nwachukwu, J.C. et al. Improved crystallographic structures using extensive combinatorial refinement. Structure 21, 1923–1930 (2013).
Debreczeni, J.E. & Emsley, P. Handling ligands with Coot. Acta Crystallogr. D Biol. Crystallogr. 68, 425–430 (2012).
McNicholas, S., Potterton, E., Wilson, K.S. & Noble, M.E. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67, 386–394 (2011).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Chalmers, M.J. et al. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 78, 1005–1014 (2006).
Busby, S.A., Chalmers, M.J. & Griffin, P.R. Improving digestion efficiency under H/D exchange conditions with activated pepsinogen coupled columns. Int. J. Mass Spectrom. 259, 130–139 (2007).
Pascal, B.D. et al. HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 23, 1512–1521 (2012).
Acknowledgements
Research support from the National Institutes of Health (PHS 5R37DK015556 to J.A.K.; 5R33CA132022, 5R01DK077085 to K.W.N.; 1U01GM102148 to K.W.N and P.R.G., and 5R01CA130932 to J.F.), The Breast Cancer Research Foundation (to B.S.K.), BallenIsles Men's Golf Association (to J.C.N.), Frenchman's Creek Women for Cancer Research (to S.S.), Susan G. Komen for the Cure (PDF12229484 to IK), and the National Natural Science Foundation of China (81172935, 81373255, 81573279), Hubei Province's Outstanding Medical Academic Leader Program (to H.-B.Z.). Use of the Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or the NIH.
Author information
Authors and Affiliations
Contributions
S.S. and J.C.N. contributed equally to this work. S.S. and J.C.N. designed and performed experiments, and wrote the manuscript; N.E.B., V.C., and J.N. performed in vitro experiments; P.R.G., S.N., V.D. and D.G. designed, performed and interpreted HDX based experiments; J.F. and I.K. contributed mammosphere assays; B.S.K., N.B. and Y.Z. designed and contributed experiments including in vivo experiments in mice; H-.B.Z., J.A.K., and J.M. designed, synthesized and performed chemical analysis of compounds; B.S.K. and J.A.K. contributed in writing and revising of the manuscript; K.W.N. designed experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Table 1 and Supplementary Figures 1–16. (PDF 4539 kb)
Rights and permissions
About this article
Cite this article
Srinivasan, S., Nwachukwu, J., Bruno, N. et al. Full antagonism of the estrogen receptor without a prototypical ligand side chain. Nat Chem Biol 13, 111–118 (2017). https://doi.org/10.1038/nchembio.2236
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2236
This article is cited by
-
Pocket similarity identifies selective estrogen receptor modulators as microtubule modulators at the taxane site
Nature Communications (2019)
-
Discovery of novel oestrogen receptor α agonists and antagonists by screening a revisited privileged structure moiety for nuclear receptors
Scientific Reports (2019)
-
A Computational Assay of Estrogen Receptor α Antagonists Reveals the Key Common Structural Traits of Drugs Effectively Fighting Refractory Breast Cancers
Scientific Reports (2018)
-
Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance
Nature Reviews Cancer (2018)
-
Specific stereochemistry of OP-1074 disrupts estrogen receptor alpha helix 12 and confers pure antiestrogenic activity
Nature Communications (2018)