Abstract
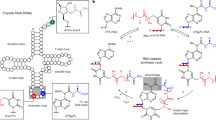
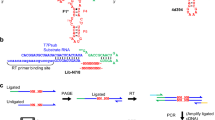
The emergence of functional interactions between nucleic acids and polypeptides was a key transition in the origin of life and remains at the heart of all biology. However, how and why simple non-coded peptides could have become critical for RNA function is unclear. Here, we show that putative ancient peptide segments from the cores of both ribosomal subunits enhance RNA polymerase ribozyme (RPR) function, as do derived homopolymeric peptides comprising lysine or the non-proteinogenic lysine analogues ornithine or, to a lesser extent, diaminobutyric acid, irrespective of chirality or chiral purity. Lysine decapeptides enhance RPR function by promoting holoenzyme assembly through primer–template docking, accelerate RPR evolution, and allow RPR-catalysed RNA synthesis at near physiological (≥1 mM) Mg2+ concentrations, enabling templated RNA synthesis within membranous protocells. Our results outline how compositionally simple, mixed-chirality peptides may have augmented the functional potential of early RNAs and promoted the emergence of the first protocells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Gesteland, R., Cech, T. & Atkins, J. (eds) The RNA World 3rd edn (Cold Spring Harbor Laboratory, 2006).
Szostak, J. W., Bartel, D. P. & Luisi, P. L. Synthesizing life. Nature 409, 387–390 (2001).
Johnston, W. K., Unrau, P. J., Lawrence, M. S., Glasner, M. E. & Bartel, D. P. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 292, 1319–1325 (2001).
Wochner, A., Attwater, J., Coulson, A. & Holliger, P. Ribozyme-catalyzed transcription of an active ribozyme. Science 332, 209–212 (2011)
Horning, D. P. & Joyce, G. F. Amplification of RNA by an RNA polymerase ribozyme. Proc. Natl Acad. Sci. USA 113, 9786–9791 (2016)
Attwater, J., Wochner, A. & Holliger, P. In-ice evolution of RNA polymerase ribozyme activity. Nat. Chem. 5, 1011–1018 (2013).
Attwater, J. et al. Chemical fidelity of an RNA polymerase ribozyme. Chem. Sci. 4, 2804–2814 (2013).
Muller, U. F. & Bartel, D. P. Improved polymerase ribozyme efficiency on hydrophobic assemblies. RNA 14, 552–562 (2008).
Attwater, J., Wochner, A., Pinheiro, V. B., Coulson, A. & Holliger, P. Ice as a protocellular medium for RNA replication. Nat. Commun. 1, 1–8 (2010).
Monnard, P. A., Apel, C. L., Kanavarioti, A. & Deamer, D. W. Influence of ionic inorganic solutes on self-assembly and polymerization processes related to early forms of life: implications for a prebiotic aqueous medium. Astrobiology 2, 139–152 (2002).
Chen, I. A., Salehi-Ashtiani, K. & Szostak, J. W. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 127, 13213–13219 (2005).
Patel, B. H., Percivalle, C., Ritson, D. J., Duffy, C. D. & Sutherland, J. D. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem. 7, 301–307 (2015).
Valadkhan, S., Mohammadi, A., Jaladat, Y. & Geisler, S. Protein-free small nuclear RNAs catalyze a two-step splicing reaction. Proc. Natl Acad. Sci. USA 106, 11901–11906 (2009).
Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000).
Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N. & Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849–857 (1983).
Coetzee, T., Herschlag, D. & Belfort, M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev. 8, 1575–1588 (1994).
Herschlag, D., Khosla, M., Tsuchihashi, Z. & Karpel, R. L. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 13, 2913–2924 (1994).
Bokov, K. & Steinberg, S. V. A hierarchical model for evolution of 23S ribosomal RNA. Nature 457, 977–980 (2009).
Hsiao, C., Mohan, S., Kalahar, B. K. & Williams, L. D. Peeling the onion: ribosomes are ancient molecular fossils. Mol. Biol. Evol. 26, 2415–2425 (2009).
Klein, D. J., Moore, P. B. & Steitz, T. A. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10, 1366–1379 (2004).
Smith, T. F., Lee, J. C., Gutell, R. R. & Hartman, H. The origin and evolution of the ribosome. Biol. Direct. 3, 16 (2008).
Alva, V., Soding, J. & Lupas, A. N. A vocabulary of ancient peptides at the origin of folded proteins. eLife 4, e09410 (2015).
Voorhees, R. M., Schmeing, T. M., Kelley, A. C. & Ramakrishnan, V. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330, 835–838 (2010).
DeRouchey, J., Hoover, B. & Rau, D. C. A comparison of DNA compaction by arginine and lysine peptides: a physical basis for arginine rich protarnines. Biochemistry 52, 3000–3009 (2013).
Wang, Q. S., Cheng, L. K. & Unrau, P. J. Characterization of the B6.61 polymerase ribozyme accessory domain. RNA 17, 469–477 (2011).
Pinheiro, V. B. et al. Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344 (2012).
Attwater, J. & Holliger, P. A synthetic approach to abiogenesis. Nat. Methods 11, 495–498 (2014).
Engelhart, A. E., Adamala, K. P. & Szostak, J. W. A simple physical mechanism enables homeostasis in primitive cells. Nat. Chem. 8, 448–453 (2016).
Adamala, K. & Szostak, J. W. Nonenzymatic template-directed RNA synthesis inside model protocells. Science 342, 1098–1100 (2013).
Kamat, N. P., Tobe, S., Hill, I. T. & Szostak, J. W. Electrostatic localization of RNA to protocell membranes by cationic hydrophobic peptides. Angew. Chem. Int. Ed. 54, 11735–11739 (2015).
Jia, T. Z., Fahrenbach, A. C., Kamat, N. P., Adamala, K. P. & Szostak, J. W. Oligoarginine peptides slow strand annealing and assist non-enzymatic RNA replication. Nat. Chem. 8, 915–921 (2016).
Cech, T. R. Evolution of biological catalysis: ribozyme to RNP enzyme. Cold Spring Harb. Symp. Quant. Biol. 74, 11–16 (2009).
Krupkin, M. et al. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos. Trans. R. Soc. Lond. B 366, 2972–2978 (2011).
Petrov, A. S. et al. History of the ribosome and the origin of translation. Proc. Natl Acad. Sci. USA 112, 15396–15401 (2015).
Hartman, H. & Smith, T. F. The evolution of the ribosome and the genetic code. Life (Basel) 4, 227–249 (2014).
Szathmary, E. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends Genet. 15, 223–229 (1999).
Johnsson, K., Allemann, R. K., Widmer, H. & Benner, S. A. Synthesis, structure and activity of artificial, rationally designed catalytic polypeptides. Nature 365, 530–532 (1993).
Muller, M. M., Windsor, M. A., Pomerantz, W. C., Gellman, S. H. & Hilvert, D. A rationally designed aldolase foldamer. Angew. Chem. Int. Ed. 48, 922–925 (2009).
Koga, S., Williams, D. S., Perriman, A. W. & Mann, S. Peptide–nucleotide microdroplets as a step towards a membrane-free protocell model. Nat. Chem. 3, 720–724 (2011).
Mann, S. Systems of creation: the emergence of life from nonliving matter. Acc. Chem. Res. 45, 2131–2141 (2012).
Thomen, P. et al. T7 RNA polymerase studied by force measurements varying cofactor concentration. Biophys. J. 95, 2423–2433 (2008).
Miele, Y., Bánsági, T. Jr, Taylor, A. F., Stano, P. & Rossi, F. in Advances in Artificial Life, Evolutionary Computation and Systems Chemistry Vol. 587 (eds Rossi, F. et al.) Ch. 18, 197–208 (Springer, 2016).
Fujii, S. et al. Liposome display for in vitro selection and evolution of membrane proteins. Nat. Protoc. 9, 1578–1591 (2014).
Acknowledgements
The authors thank S. James for assistance with fidelity data processing. This work was supported by postdoctoral fellowships from JSPS (Japanese Society for the Promotion of Science) and HFSP (Human Frontiers Science Program) (S.T.) and by the Medical Research Council (J.A., P.H.; programme no. MC_U105178804).
Author information
Authors and Affiliations
Contributions
S.T. and P.H. conceived and designed the experiments. S.T. performed all experiments together with J.A. for selection design, fidelity measurement and peptide assays. All authors discussed the results and jointly wrote and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 11883 kb)
Rights and permissions
About this article
Cite this article
Tagami, S., Attwater, J. & Holliger, P. Simple peptides derived from the ribosomal core potentiate RNA polymerase ribozyme function. Nature Chem 9, 325–332 (2017). https://doi.org/10.1038/nchem.2739
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2739
This article is cited by
-
Boron-assisted abiotic polypeptide synthesis
Communications Chemistry (2023)
-
Hydrophobic-cationic peptides modulate RNA polymerase ribozyme activity by accretion
Nature Communications (2022)
-
The Origin(s) of Cell(s): Pre-Darwinian Evolution from FUCAs to LUCA
Journal of Molecular Evolution (2021)
-
Mutually stabilizing interactions between proto-peptides and RNA
Nature Communications (2020)
-
Template-directed RNA polymerization and enhanced ribozyme catalysis inside membraneless compartments formed by coacervates
Nature Communications (2019)