Abstract
Although genetic factors contribute to almost half of all cases of deafness, treatment options for genetic deafness are limited1,2,3,4,5. We developed a genome-editing approach to target a dominantly inherited form of genetic deafness. Here we show that cationic lipid-mediated in vivo delivery of Cas9–guide RNA complexes can ameliorate hearing loss in a mouse model of human genetic deafness. We designed and validated, both in vitro and in primary fibroblasts, genome editing agents that preferentially disrupt the dominant deafness-associated allele in the Tmc1 (transmembrane channel-like gene family 1) Beethoven (Bth) mouse model, even though the mutant Tmc1Bth allele differs from the wild-type allele at only a single base pair. Injection of Cas9–guide RNA–lipid complexes targeting the Tmc1Bth allele into the cochlea of neonatal Tmc1Bth/+ mice substantially reduced progressive hearing loss. We observed higher hair cell survival rates and lower auditory brainstem response thresholds in injected ears than in uninjected ears or ears injected with control complexes that targeted an unrelated gene. Enhanced acoustic startle responses were observed among injected compared to uninjected Tmc1Bth/+ mice. These findings suggest that protein–RNA complex delivery of target gene-disrupting agents in vivo is a potential strategy for the treatment of some types of autosomal-dominant hearing loss.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Angeli, S., Lin, X. & Liu, X. Z. Genetics of hearing and deafness. Anat. Rec. (Hoboken) 295, 1812–1829 (2012)
Marazita, M. L. et al. Genetic epidemiological studies of early-onset deafness in the U.S. school-age population. Am. J. Med. Genet. 46, 486–491 (1993)
Morton, C. C. & Nance, W. E. Newborn hearing screening—a silent revolution. N. Engl. J. Med. 354, 2151–2164 (2006)
Géléoc, G. S. & Holt, J. R. Sound strategies for hearing restoration. Science 344, 1241062 (2014)
Müller, U. & Barr-Gillespie, P. G. New treatment options for hearing loss. Nat. Rev. Drug Discov. 14, 346–365 (2015)
Shibata, S. B. et al. RNA interference prevents autosomal-dominant hearing loss. Am. J. Hum. Genet. 98, 1101–1113 (2016)
Pan, B. et al. Gene therapy restores auditory and vestibular function in a mouse model of Usher syndrome type 1c. Nat. Biotechnol. 35, 264–272 (2017)
Ahmed H., Shubina-Oleinik O. & Holt, J. R. Emerging gene therapies for genetic hearing loss. J. Assoc. Res. Otolaryngol. 18, 649–670 (2017)
Sacheli, R., Delacroix, L., Vandenackerveken, P., Nguyen, L. & Malgrange, B. Gene transfer in inner ear cells: a challenging race. Gene Ther. 20, 237–247 (2013)
Komor, A. C., Badran, A. H. & Liu, D. R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 (2017)
Long, C. et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403 (2016)
Yang, Y. et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 34, 334–338 (2016)
Yin, H. et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 34, 328–333 (2016)
Kim, S., Kim, D., Cho, S. W., Kim, J. & Kim, J. S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014)
Zuris, J. A. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015)
Pan, B. et al. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79, 504–515 (2013)
Camp, G. V. & Smith, R. Hereditary Hearing Loss Homepagehttp://hereditaryhearingloss.org (2017)
Zhao, Y. et al. A novel DFNA36 mutation in TMC1 orthologous to the Beethoven (Bth) mouse associated with autosomal dominant hearing loss in a Chinese family. PLoS One 9, e97064 (2014)
Kurima, K. et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat. Genet. 30, 277–284 (2002)
Kawashima, Y., Kurima, K., Pan, B., Griffith, A. J. & Holt, J. R. Transmembrane channel-like (TMC) genes are required for auditory and vestibular mechanosensation. Pflugers Arch. 467, 85–94 (2015)
Vreugde, S. et al. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat. Genet. 30, 257–258 (2002)
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M. & Joung, J. K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284 (2014)
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015)
Zhang, F. CRISPR Designhttp://crispr.mit.edu (2015)
Lentz, J. J. et al. Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat. Med. 19, 345–350 (2013)
Wang, M. et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl Acad. Sci. USA 113, 2868–2873 (2016)
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843 (2013)
Lumpkin, E. A. et al. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns 3, 389–395 (2003)
Huang, M., Kantardzhieva, A., Scheffer, D., Liberman, M. C. & Chen, Z. Y. Hair cell overexpression of Islet1 reduces age-related and noise-induced hearing loss. J. Neurosci. 33, 15086–15094 (2013)
Acknowledgements
This work was supported by DARPA HR0011-17-2-0049 (to D.R.L.), US NIH R01 EB022376 (to D.R.L.), R35 GM118062 (to D.R.L.), R01 DC006908 (to Z.-Y.C), P30 DC05209 (to M.C.L.), R01 DC00138 (to M.C.L.), and R01 DC013521 (to J.R.H.). We are grateful for support from the David-Shulsky Foundation (to Z.-Y.C.), a Frederick and Ines Yeatts Hair Cell Regeneration grant (to Y.T., V.L., M.H., and Y.S.), the Bertarelli Foundation and the Jeff and Kimberly Barber Fund (to J.R.H.), the Broad Institute (to D.R.L and Z.-Y.C.), and the HHMI (to D.R.L.). We thank H. Rees, S. Tsai, M. Packer, K. Zhao and D. Usanov for assistance.
Author information
Authors and Affiliations
Contributions
X.G. and Y.T. designed the research, performed genome editing (X.G.) and hearing biology (Y.T.) experiments, analysed data, and wrote the manuscript. V.L., M.H., W.-H.Y., B.P., Y.-J.H., and H.W. designed experiments, performed hearing biology experiments and analysed data. D.B.P., M.C.L., and W.-J.K. designed hearing experiments and analysed data. Y.S. and S.Y. supported hearing biology experiments. J.H.H. analysed GUIDE-seq data. D.B.T. supported genome editing experiments. Y.L. and Q.X. designed and synthesized lipids. J.R.H., Z.-Y.C. and D.R.L. designed and supervised the research, and wrote the manuscript. All authors edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
D.R.L. is a consultant and co-founder of Editas Medicine, Beam Therapeutics, and Pairwise Plants, companies that use genome editing. X.G., D.B.T., Z.-Y.C., and D.R.L. have filed patent applications on aspects of this work.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
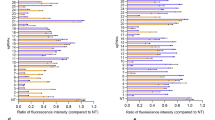
Extended Data Figure 1 Allele-selective editing of wild-type or Bth mutant Tmc1 in cleavage assays in vitro and by lipid-mediated delivery into primary fibroblasts.
a, In vitro Cas9–sgRNA-mediated Tmc1 DNA cleavage. We incubated 100 nM of a 995-bp DNA fragment containing wild-type Tmc1 (lanes 1–5) or Tmc1Bth (lanes 6–10) with 300 nM of each of the four Cas9–sgRNAs shown for 15 min at 37 °C. Expected cleavage products are 774–778 bp and 217–221 bp. M, 100-bp ladder; the lower two heavy bands are 500 and 1,000 bp. b, Quantification of DNA cleavage in a by densitometry using imageJ. c, Comparison of transfection efficiency in HEK293T cells and wild-type primary fibroblasts. Fifty nanograms GFP plasmid, 10 nM Cas9–FitC–Tmc1-mut3 sgRNA RNP, or 10 nM Cas9–CrRNA–Tmc1-mut3–atto-550-TracrRNA RNP were delivered into HEK293T cells or wild-type primary fibroblasts using 3 μl Lipofectamine 2000. For samples with GFP plasmid, the fraction of GFP-positive cells was measured by flow cytometry 24 h after delivery. For samples with Cas9–FitC–Tmc1-mut3 RNP or Cas9–CrRNA–Tmc1-mut3–atto-550-TracrRNA RNP, medium was removed 6 h after delivery. The cells were trypsinized, washed three times with 500 μl PBS containing 20 U ml−1 heparin, and subjected to flow cytometry. d, Wild-type or Bth mutant Tmc1 allele editing in primary fibroblasts derived from wild-type or Tmc1Bth/Bth mice as a function of the dose of Cas9–Tmc1-mut3–lipid complex. Cas9–Tmc1-mut3 (12.5, 25, 50, 100, 200, or 400 nM) was delivered into the primary fibroblasts using Lipofectamine 2000 in DMEM–FBS. e, Lipid-mediated delivery of Cas9–sgRNA complexes into primary fibroblasts derived from wild-type or Tmc1Bth/Bth mice. We delivered 100 nM of purified Cas9 protein and each wild-type Tmc1-targeting sgRNA (Tmc1-wt1, Tmc1-wt2, or Tmc1-wt3) or Tmc1Bth mutant-targeting sgRNA (Tmc1-mut1, Tmc1-mut2, or Tmc1-mut3) into wild-type fibroblasts (red) and Tmc1Bth/Bth fibroblasts (blue) using Lipofectamine 2000 in DMEM–FBS. Primary fibroblast cells were harvested 96 h after treatment. Genomic DNA was extracted and indels were detected by HTS. Individual values (n = 3–4) are shown; horizontal lines and error bars represent mean ± s.d. of biological replicates.
Extended Data Figure 2 Delivery of Cas9–Tmc1-mut3 sgRNA complexes into primary fibroblasts derived from wild-type or homozygous Tmc1Bth/Bth mice.
a, Using seven commercially available lipids: LPF2000 (Lipofectamine 2000); RNAiMAX (Lipofectamine RNAiMAX); LPF3000 (Lipofectamine 3000); CRISPRMAX (Lipofectamine CRISPRMAX); LTX (Lipofectamine LTX). b, Using ten biodegradable, bioreducible lipids: Lipid 1 (75-O14B); Lipid 2 (76-O14B); Lipid 3 (80-O18B); Lipid 4 (87-O16B); Lipid 5 (113-O18B); Lipid 6 (306-O12B); Lipid 7 (306-O16B); Lipid 8 (306-O18B); Lipid 9 (400-O12B); Lipid 10 (400-O16B). We delivered 100 nM purified Cas9–Tmc1-mut3 RNP using 3 μl of the cationic lipid shown in DMEM-FBS. Fibroblast cells were collected 96 h after treatment, genomic DNA was extracted, and indels were detected by HTS. c, Synthetic route and chemical structure of lipids. d, Commercially available amine head groups used in lipid synthesis. Lipids were synthesized as previously described26. Individual values (n = 2–4) are shown; horizontal lines and error bars represent mean ± s.d. of three or more biological replicates.
Extended Data Figure 3 Off-target sites identified by GUIDE-seq after nucleofection of DNA plasmids encoding Cas9 and Tmc1-mut3 sgRNA into primary fibroblasts from Tmc1Bth/+ mice.
a, One thousand nanograms of Cas9 plasmid, 300 ng Tmc1-mut3 sgRNA plasmid, 400 ng pmaxGFP plasmid, and 50 pmol double-stranded oligodeoxynucleotides (dsODN) were nucleofected into Tmc1Bth/+ fibroblasts using a LONZA 4D-Nucleofector. Genomic DNA was extracted 96 h after nucleofection and subjected to GUIDE-seq as previously described23. Off-T1 to Off-T10 are ten off-target sites detected by GUIDE-seq. Mismatches compared to the on-target site are shown and highlighted in colour. The Tmc1Bth allele targeted by sgRNA Tmc1-mut3 is shown in the top row. b, Indel frequency at the Tmc1 locus and at each of the off-target loci in Cas9–Tmc1-mut3-treated Tmc1Bth/Bth primary fibroblasts following plasmid DNA nucleofection or following RNP delivery. For RNP delivery, 100 nM Cas9–Tmc1-mut3 RNP was delivered to the Tmc1Bth/Bth fibroblasts using 3 μl Lipofectamine 2000. Indels were detected by HTS at the Tmc1 on-target site and at each off-target site. Red, samples nucleofected with DNA plasmids encoding Cas9 and Tmc1-mut3 sgRNA; blue, samples treated with Cas9–Tmc1-mut3 RNPs; grey, control samples nucleofected with unrelated dsDNA only.
Extended Data Figure 4 Cas9–Tmc1-mut3–lipid injection reduces hearing loss, improves acoustic startle response, and preserves stereocilia in Tmc1Bth/+ mice.
a, Phalloidin labelling showed the preservation of stereocilia of IHCs in an ear eight weeks after injection with Cas9–Tmc1-mut3 sgRNA at three frequency locations indicated, whereas the uninjected contralateral inner ear of the same mouse showed severe degeneration of stereocilia at locations corresponding to 16 and 32 kHz. The boxes indicate the stereocilia, which are shown at the bottom of each image at higher magnification. Scale bars, 10 μm. Similar results were observed in other injected ears that were immunolabelled (n = 5). b, Representative ABR waveforms showing reduced threshold (red traces) at 16 kHz in a Cas9–Tmc1-mut3–lipid-injected Tmc1Bth/+ ear (left) compared to the uninjected contralateral ear (right) of the same mouse after four weeks. c, Eight weeks after Cas9–Tmc1-mut3 injection into Tmc1Bth/+ ears (blue), mean ABR thresholds were significantly reduced at three frequencies. Uninjected Tmc1Bth/+ ears (red) showed ABR thresholds >85 dB at all frequencies after eight weeks. ABR thresholds from wild-type C3H mice are shown in green. d, ABR wave 1 amplitudes following 90 dB SPL stimulation at 16 kHz were greater in injected Tmc1Bth/+ ears than in uninjected ears eight weeks after treatment. Individual values (n = 15 or 20 for uninjected, and 24 for injected) are shown; horizontal bars represent mean values. e, Startle responses at 16 kHz in individual Cas9–Tmc1-mut3 sgRNA-injected mice (blue) were significantly stronger (P < 0.001) than in uninjected mice (red) eight weeks after treatment. Among the different frequencies assayed, the number of ears tested (n) varies within the range shown (Supplementary Table 2). Statistical analyses of ABR thresholds, amplitudes, and startle responses were performed by two-way ANOVA with Bonferroni correction for multiple comparisons: *P < 0.05, **P < 0.01, ****P < 0.0001. Values and error bars reflect mean ± s.e.m.
Extended Data Figure 5 Effect of in vivo injection of Cas9–sgRNA–lipid complexes on DPOAE thresholds.
a–d, DPOAE thresholds four weeks after injection were elevated compared with uninjected ears at three frequencies following treatment with Cas9–Tmc1-mut3 sgRNA (a), and were elevated at two frequencies following treatment with Cas9–Tmc1-wt3 sgRNA (b), Cas9–GFP sgRNA (c), or dCas9–Tmc1-mut1 sgRNA (d). e, Eight weeks after Cas9–Tmc1-mut3 sgRNA injection, DPOAE thresholds were elevated at three frequencies in the injected group. Mean DPOAE thresholds of untreated wild-type (WT) C3H mice at four weeks (a) or eight weeks (e) of age are also shown in purple. Statistical analysis of DPOAE thresholds was performed by two-way ANOVA with Bonferroni correction for multiple comparisons: **P < 0.01, ****P < 0.0001. Values and error bars reflect mean ± s.e.m. Among the different frequencies assayed, the number of ears tested (n) varies within the range shown (Supplementary Table 2). The elevation of DPOAE thresholds despite enhanced hair cell survival (Fig. 2d, g) suggests that the surviving OHCs may not be fully functional. IHCs can respond to sound and excite auditory nerve fibres in the absence of OHC amplification, although at higher SPLs. Thus, an improvement in ABR thresholds and suprathreshold amplitudes can occur without concomitant DPOAE enhancement if the functional improvements are restricted to the surviving IHCs.
Extended Data Figure 6 Hearing rescue is dependent on the Tmc1Bth target specificity of the sgRNA, Cas9 nuclease activity, the presence of the Tmc1Bth mutation, and the presence of the sgRNA.
a, In Tmc1Bth/+ ears injected with Cas9–Tmc1-wt3–lipid, which targets the wild-type Tmc1 allele instead of the mutant Tmc1Bth allele, ABR thresholds (blue) were comparable to or higher than those of uninjected controls (red) after four weeks. b, Tmc1Bth/+ ears injected with Cas9–GFP sgRNA–lipid (blue) did not show improved ABR thresholds four weeks after treatment. c, Tmc1Bth/+ ears injected with catalytically inactive dCas9–Tmc1-mut1–lipid did not show improved ABR thresholds four weeks after treatment. d, ABR thresholds of wild-type C3H mice injected with Cas9–Tmc1-mut3–lipid showed similar patterns to the uninjected control inner ears at four weeks, except at 5.66 and 45.24 kHz where ABR thresholds were elevated. e, Elevated DPOAE thresholds at three frequencies were observed after the treatment in d. f, Injection of Cas9–Lipofectamine 2000 (LPF2000) without sgRNA in Tmc1Bth/+ mice did not improve ABR thresholds after four weeks. g, Elevated DPOAE thresholds at 11 and 16 kHz were observed after the treatment in f. Statistical analysis of ABR and DPOAE thresholds was performed by two-way ANOVA with Bonferroni correction for multiple comparisons: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Values and error bars reflect mean ± s.e.m. Among the different frequencies assayed, the number of ears tested (n) varies within the range shown (Supplementary Table 2).
Extended Data Figure 7 Hearing preservation following treatment with additional Tmc1-mut sgRNAs other than Tmc1-mut3.
a, Mean ABR thresholds were significantly reduced at three frequencies in ears injected with Cas9–Tmc1-mut1–lipid compared to uninjected Tmc1Bth/+ ears after four weeks. b, DPOAE thresholds were elevated in the same group of inner ears after Cas9–Tmc1-mut1 injection as in a after four weeks. c, Mean ABR thresholds were significantly reduced at five frequencies in ears injected with Cas9–Tmc1-mut2–lipid compared to uninjected Tmc1Bth/+ ears after four weeks. d, DPOAE thresholds were elevated in the same group of inner ears after Cas9–Tmc1-mut2 injection as in c after four weeks. e, Mean ABR thresholds were significantly reduced at three frequencies in ears injected with Cas9–Tmc1-mut4–lipid compared to uninjected Tmc1Bth/+ ears after four weeks. f, DPOAE thresholds were elevated in the same group of inner ears after Cas9–Tmc1-mut4–lipid injection as in e after four weeks. g, Significantly stronger wave 1 amplitudes were detected in ears injected with each of the Cas9–Tmc1-mut–lipid complexes shown at 16 kHz (80 and 90 dB SPL). Individual values (n = 8, 13, or 18) are shown; horizontal bars represent mean values. h, Eight weeks after Cas9–Tmc1-mut1–lipid injection into Tmc1Bth/+ ears, mean ABR thresholds were significantly reduced at five frequencies compared to uninjected Tmc1Bth/+ ears, which showed ABR thresholds >80 dB at all frequencies after eight weeks. Mean ABR thresholds of untreated wild-type (WT) C3H mice of eight weeks of age are shown in purple. Red arrows indicate no ABR response at the highest SPL level of 90 dB. i, DPOAE thresholds were significantly elevated at two frequencies (8 and 11 kHz) in the same group of inner ears after Cas9–Tmc1-mut1 injection as in h after eight weeks. Mean DPOAE thresholds of untreated wild-type C3H mice of eight weeks of age are shown in purple. Statistical analysis of ABR and DPOAE thresholds and wave 1 amplitudes was performed by two-way ANOVA with Bonferroni correction for multiple comparisons: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Values and error bars reflect mean ± s.e.m. Among the different frequencies assayed, the number of ears tested (n) varies within the range shown (Supplementary Table 2).
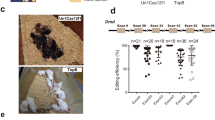
Extended Data Figure 8 RNP delivery of Cas9–sgRNA–lipid complexes results in genome editing in adult hair cells.
Six-week-old adult Atoh1–GFP cochlea were injected with 1 μl 25 μM Cas9–GFP sgRNA–lipid complex by canalostomy, with the cochlea removed two weeks after injection. a, Genome editing was detected by the loss of GFP (green, with GFP absence noted using cyan shapes) in inner hair cells (IHCs) and outer hair cells (OHCs). b, Hair cells were labelled with the hair cell marker MYO7A (red) in the apex turn of cochlea. c, d, In uninjected contralateral Atoh1–GFP cochlea, all hair cells were GFP-positive. Scale bars, 10 μm. Similar results were observed in other injected ears that were immunolabelled (n = 3).
Extended Data Figure 9 In vivo editing of the Tmc1 locus from Tmc1Bth/+ ears injected with Cas9–Tmc1-mut3 sgRNA.
A representation of the organ of Corti removed at P5 for high-throughput DNA sequencing. a, A confocal z-stack image showing the surface view of a dissected and labelled organ of Corti used for HTS. b, A cross-sectional view of the organ of Corti (along the white line in a) showing the positions of hair cells (MYO7A), supporting cells (SOX2) and the cells from other cochlear regions that were used for quantification. LER, lesser epithelial ridge; GER, greater epithelial ridge; SE, sensory epithelium; Lib, limbus region. DAPI-labelled nuclei are shown in blue. Quantification showed that hair cells represented 1.45% ± 0.05% (mean ± s.e.m., n = 4) of all cells in the dissected cochlea. Scale bars, 10 μm. c, On-target and off-target in vivo editing of the Tmc1 locus in organ of Corti samples. No indels were observed at frequencies substantially above that of an untreated control sample at any of the ten off-target sites identified by GUIDE-seq (Off-T1 to Off-T10). Indels were detected by HTS at the Tmc1 on-target site and each off-target site from in vivo tissue samples dissected from the inner ear of neonatal mice 4 days after Cas9–Tmc1-mut3 RNP injection (blue), or from untreated control samples (red).
Supplementary information
Supplementary Information
This file contains a representative example of the gating strategy used in the flow cytometry experiments in Extended Data Figure 1a. (PDF 335 kb)
Supplementary Information
This file contains supplementary methods which includes tables 1 and 2, sequences, notes and figure 1. (PDF 593 kb)
Rights and permissions
About this article
Cite this article
Gao, X., Tao, Y., Lamas, V. et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature 553, 217–221 (2018). https://doi.org/10.1038/nature25164
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25164
This article is cited by
-
Genetic correction of induced pluripotent stem cells from a DFNA36 patient results in morphologic and functional recovery of derived hair cell-like cells
Stem Cell Research & Therapy (2024)
-
Distributional comparison of different AAV vectors after unilateral cochlear administration
Gene Therapy (2024)
-
Strategies for non-viral vectors targeting organs beyond the liver
Nature Nanotechnology (2024)
-
An Update on the Application of CRISPR Technology in Clinical Practice
Molecular Biotechnology (2024)
-
Novel autosomal dominant TMC1 variants linked to hearing loss: insight into protein-lipid interactions
BMC Medical Genomics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.