Abstract
Fibrosis is a common pathology in cardiovascular disease1. In the heart, fibrosis causes mechanical and electrical dysfunction1,2 and in the kidney, it predicts the onset of renal failure3. Transforming growth factor β1 (TGFβ1) is the principal pro-fibrotic factor4,5, but its inhibition is associated with side effects due to its pleiotropic roles6,7. We hypothesized that downstream effectors of TGFβ1 in fibroblasts could be attractive therapeutic targets and lack upstream toxicity. Here we show, using integrated imaging–genomics analyses of primary human fibroblasts, that upregulation of interleukin-11 (IL-11) is the dominant transcriptional response to TGFβ1 exposure and required for its pro-fibrotic effect. IL-11 and its receptor (IL11RA) are expressed specifically in fibroblasts, in which they drive non-canonical, ERK-dependent autocrine signalling that is required for fibrogenic protein synthesis. In mice, fibroblast-specific Il11 transgene expression or Il-11 injection causes heart and kidney fibrosis and organ failure, whereas genetic deletion of Il11ra1 protects against disease. Therefore, inhibition of IL-11 prevents fibroblast activation across organs and species in response to a range of important pro-fibrotic stimuli. These results reveal a central role of IL-11 in fibrosis and we propose that inhibition of IL-11 is a potential therapeutic strategy to treat fibrotic diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Rockey, D. C., Bell, P. D. & Hill, J. A. Fibrosis—a common pathway to organ injury and failure. N. Engl. J. Med. 372, 1138–1149 (2015)
Burstein, B. & Nattel, S. Atrial fibrosis: mechanisms and clinical relevance in atrial fibrillation. J. Am. Coll. Cardiol. 51, 802–809 (2008)
Leaf, I. A. & Duffield, J. S. What can target kidney fibrosis? Nephrol. Dial. Transplant. 32 (suppl. 1), i89–i97 (2017)
Davis, J. & Molkentin, J. D. Myofibroblasts: trust your heart and let fate decide. J. Mol. Cell. Cardiol. 70, 9–18 (2014)
Akhurst, R. J. & Hata, A. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 11, 790–811 (2012)
Bierie, B. et al. Abrogation of TGF-β signaling enhances chemokine production and correlates with prognosis in human breast cancer. J. Clin. Invest. 119, 1571–1582 (2009)
Shull, M. M. et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature 359, 693–699 (1992)
Wynn, T. A. Cellular and molecular mechanisms of fibrosis. J. Pathol. 214, 199–210 (2008)
Cucoranu, I. et al. NAD(P)H oxidase 4 mediates transforming growth factor-β1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circ. Res. 97, 900–907 (2005)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013)
FANTOM Consortium and the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014)
Schmitt, J. P. et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science 299, 1410–1413 (2003)
Du, X. X., Neben, T., Goldman, S. & Williams, D. A. Effects of recombinant human interleukin-11 on hematopoietic reconstitution in transplant mice: acceleration of recovery of peripheral blood neutrophils and platelets. Blood 81, 27–34 (1993)
Putoczki, T. L. et al. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell 24, 257–271 (2013)
Obana, M. et al. Therapeutic activation of signal transducer and activator of transcription 3 by interleukin-11 ameliorates cardiac fibrosis after myocardial infarction. Circulation 121, 684–691 (2010)
Ernst, M. & Putoczki, T. L. Molecular pathways: IL11 as a tumor-promoting cytokine—translational implications for cancers. Clin. Cancer Res. 20, 5579–5588 (2014)
Lokau, J. et al. Proteolytic cleavage governs interleukin-11 trans-signaling. Cell Reports 14, 1761–1773 (2016)
Dams-Kozlowska, H. et al. A designer hyper interleukin 11 (H11) is a biologically active cytokine. BMC Biotechnol. 12, 8 (2012)
Nandurkar, H. H. et al. Adult mice with targeted mutation of the interleukin-11 receptor (IL11Ra) display normal hematopoiesis. Blood 90, 2148–2159 (1997)
Duan, J. et al. Wnt1/βcatenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. EMBO J. 31, 429–442 (2012)
Kaye, J. A. FDA licensure of NEUMEGA to prevent severe chemotherapy-induced thrombocytopenia. Stem Cells 16 (Suppl 2), 207–223 (1998)
Moore-Morris, T. et al. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J. Clin. Invest. 124, 2921–2934 (2014)
Paul, S. R. et al. Molecular cloning of a cDNA encoding interleukin 11, a stromal cell-derived lymphopoietic and hematopoietic cytokine. Proc. Natl Acad. Sci. USA 87, 7512–7516 (1990)
Nakagawa, M. et al. Four cases of investigational therapy with interleukin-11 against acute myocardial infarction. Heart Vessels 31, 1574–1578 (2016)
Lindahl, G. E. et al. Microarray profiling reveals suppressed interferon stimulated gene program in fibroblasts from scleroderma-associated interstitial lung disease. Respir. Res. 14, 80 (2013)
Nieminen, P. et al. Inactivation of IL11 signaling causes craniosynostosis, delayed tooth eruption, and supernumerary teeth. Am. J. Hum. Genet. 89, 67–81 (2011)
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev. A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015)
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009)
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015)
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005)
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008)
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014)
Babraham Bioinformatics. FastQC, a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014)
Burke, M. A. et al. Molecular profiling of dilated cardiomyopathy that progresses to heart failure. JCI Insight 1, e86898 (2016)
Tarnavski, O. et al. Mouse cardiac surgery: comprehensive techniques for the generation of mouse models of human diseases and their application for genomic studies. Physiol. Genomics 16, 349–360 (2004)
Zheng, B., Zhang, Z., Black, C. M., de Crombrugghe, B. & Denton, C. P. Ligand-dependent genetic recombination in fibroblasts : a potentially powerful technique for investigating gene function in fibrosis. Am. J. Pathol. 160, 1609–1617 (2002)
Ye, L. et al. Thymosin β4 increases the potency of transplanted mesenchymal stem cells for myocardial repair. Circulation 128 (Suppl 1), S32–S41 (2013)
Gao, S., Ho, D., Vatner, D. E. & Vatner, S. F. Echocardiography in mice. Curr. Protoc. Mouse Biol. 1, 71–83 (2011)
Tortoledo, F. A., Quinones, M. A., Fernandez, G. C., Waggoner, A. D. & Winters, W. L. Jr. Quantification of left ventricular volumes by two-dimensional echocardiography: a simplified and accurate approach. Circulation 67, 579–584 (1983)
Mead, T. J. & Lefebvre, V. Proliferation assays (BrdU and EdU) on skeletal tissue sections. Methods Mol. Biol. 1130, 233–243 (2014)
Acknowledgements
We thank all patients for taking part in this research, which was performed with approval from the SingHealth Centralised IRB Review Board (CIRB; 2013/103/C). The research was supported by the National Medical Research Council (NMRC) Singapore STaR award (S.A.C.) (NMRC/STaR/0011/2012), the NMRC Centre Grant to the National Heart Centre Singapore (NHCS), Goh Foundation, Tanoto Foundation, NHLBI 5R01HL080494 (J.G.S., C.E.S.), HHMI (C.E.S.) and a grant from the Fondation Leducq (N.H., J.G.S., C.E.S., S.C.). We thank I. Kamer and R. Plehm, Max-Delbrück-Center for Molecular Medicine (MDC), for expert technical help with telemetry blood pressure measurements.
Author information
Authors and Affiliations
Contributions
S.A.C. conceived, managed and arranged funding for the project. Wet lab experiments (cell culture, cell biology, molecular biology, RNA-seq) were carried out by S.V., A.A.W., W.-W.L., B.N., G.P., J.T., L.Y., N.E.S., C.J.P., C.X., M.W., S.L., K.Sa., S.B., S.Schm., T.P., N.G.-C., H.W., S.v.H. and K.Si. Single-cell studies were carried out by D.M.D., J.G.S. and C.E.S. In vivo gain-of-function and loss-of-function mouse experiments were performed by A.A.W., W.-W.L., B.N., J.T., E.K., L.Y., N.S.J.K., N.T.G.Z., D.M.D., G.P. and H.M. Data were analysed by S.Scha., S.V., A.A.W., A.M.-M., K.C., S.P.C., O.J.L.R., K.Sa., E.P., S.M.E., J.G.S., C.E.S. and N.H. Patient-based studies were carried out by S.L.L., J.L.S., V.T.T.C., Y.L.C., T.E.T., Y.J.L., M.H.J., K.K.O., K.C.C., B.-H.O., M.J.C. and K.Y.K.S. S.Scha., S.V., A.A.W. and S.A.C. designed experiments and prepared the manuscript with input from co-authors.
Corresponding author
Ethics declarations
Competing interests
S.A.C. and S.Scha. are co-inventors of the patent application (WO2017103108) ‘Treatment of fibrosis’. S.A.C. and S.Scha. are co-founders and shareholders of Enleofen Bio PTE LTD, a company that develops therapeutics based on findings described in this manuscript.
Additional information
Reviewer Information Nature thanks S. Friedman and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
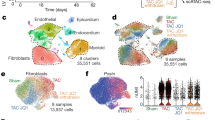
Extended Data Figure 1 RNA-seq of fibrotic cardiac fibroblasts.
a, Mapped RNA-seq reads for human fibroblasts (n = 84 biologically independent samples). We aimed to generate at least 60 million reads per sample to assess RNA expression with and without stimulation of primary cardiac fibroblasts with TGFβ1. Fibroblasts were derived from the atria of 84 individuals, resulting in a total of 168 RNA-seq datasets for unstimulated and stimulated cells. After mapping, we used only reads that map to one unique location in the genome to estimate gene expression levels. b, Distribution of mean gene expression across all samples for each gene. We found 12,081 genes to be expressed at log2(TPM + 1) > 2. Genes below this cut-off were not considered in subsequent analyses. c, TPM distribution for each sample after filtering (n = 84). d, Ward clustering of Euclidian distance of RNA-seq samples. Sample gene expression tends to cluster mostly by genotype, indicating a strong genetic effect, beyond the effects of TGFβ1 stimulation.
Extended Data Figure 2 Characterization the TGFβ1-regulated gene, Il-11.
a, We measured the amount of activated fibroblasts (ACTA2+ cells) using the Operetta High-content imaging platform and IL-11 transcript levels by RNA-seq (n = 84 biologically independent samples). The increase in IL-11 expression correlated strongly with fibroblast activation in the cohort (ρ = 0.47, 95% confidence interval = 0.28–0.62). ST, stimulated cells (5 ng ml−1, 24 h); BL, baseline cells (unstimulated, 24 h). b, TGFβ1 (5 ng ml−1, 24 h) significantly upregulates IL11 RNA (8.5-fold, P = 6 × 10−218) in human atrial fibroblasts according to RNA-seq analysis (n = 84 biologically independent samples). P values were calculated with DEseq2. c, The change in gene expression was confirmed at the protein level using an ELISA assay to measure IL-11 protein in the supernatant of cardiac fibroblasts (n = 6 biologically independent samples). fc, fold change. Two-tailed Student’s t-test. d, RNA-seq-based expression differences in IL11 transcript between unstimulated and TGFβ1-stimulated cardiac fibroblasts were confirmed via RT–qPCR. e, f, The JSD was calculated for all protein-coding genes expressed in activated fibroblasts. Low JSD scores indicate that a gene is highly expressed in stimulated cardiac fibroblasts and lowly expressed in healthy tissues (e; GTEx) or unstimulated, primary cell lines (f; FANTOM). g, h, IL11 RNA expression in TGFβ1-stimulated and unstimulated cardiac fibroblasts (n = 84 biologically independent samples) compared to GTEx (g; tissues from n = 8,723 biologically independent samples) and FANTOM (h; cell types from n = 285 biologically independent samples) databases. g, RNA expression of IL11 in TGFβ1-stimulated cardiac fibroblasts (red) and healthy tissues. h, RNA expression of IL11 in TGFβ1-stimulated cardiac fibroblasts (red) and primary cells. b, c, g, h, Box-and-whisker plots show median (middle line), 25th–75th percentiles (box) and 10th–90th percentiles (whiskers).
Extended Data Figure 3 Single-cell RNA-seq of fibrotic mouse heart.
Single-cell RNA-seq analysis of cells isolated from PlnR9C/+ and wild-type adult left ventricles shown in Fig. 1. a, Cell types were defined according to indicated marker genes. b, Cells were clustered and cell type was determined using the Seurat R package (see Methods). c, Upregulation of Il-11 in the heart in the PlnR9C/+ fibrosis model was confirmed using western blotting. All mice were 18 weeks old and male. d, Subsequently, 1,263 fibroblasts from PlnR9C/+ and wild-type mice were re-clustered using Seurat. WNT signalling, downstream of TGFβ1 in cardiac fibroblast activation, target genes were used in this analysis of the four subsequent clusters of fibroblasts; Il-11+ cells were primarily found in clusters 0 and 1 (16 out of 18 Il-11-expressing cells). Clusters 0 and 1 were also enriched for PlnR9C/+ cells compared to wild-type. Il11ra1 was expressed in all clusters. Cardiac cells were sequenced from n = 1 mouse, experiment was repeated one time with similar results.
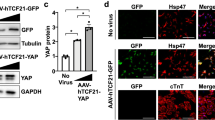
Extended Data Figure 4 IL-11 activates fibroblasts and is required for the pro-fibrotic effect of TGFβ1.
a, b, High-resolution fluorescence imaging after TGFβ1 or IL-11 treatment (5 ng ml−1, 24 h) of primary cardiac fibroblasts. Immunostaining of nuclei (DAPI, blue), ACTA2 (red) and F-actin (phalloidin, green) indicated that both TGFβ1 and IL-11 activate fibroblast stress fibre formation and increase the number of myofibroblasts in vitro to similar levels. Experiment was repeated four times with similar results. c, Automated quantification of fluorescence (Operetta assay n = 7 measurements per n = 6 independent experiments) of primary atrial fibroblasts reveals significant fibroblast activation and ECM production induced by both TGFβ1 and IL-11 (5 ng ml−1, 24 h). d, In addition, TGFβ1 effects can be reduced with an anti-IL-11 antibody (2 μg ml−1). c, d, Collagen secretion in the supernatant (n = 6 independent experiments) was assessed with Sirius Red. e, Mouse primary fibroblasts were incubated for 24 h with indicated concentrations of recombinant human or mouse IL-11. Fibroblast activation was monitored using the Operetta High-Content Imaging platform and immunostaining for ACTA2. rhIL-11 was found to inefficiently activate mouse fibroblasts (rmIL-11, n = 2, rhIL-11, n = 4 biologically independent samples) compared to rmIL-11; this occurred for rhIL-11 treatment with rhIL-11 from two separate suppliers. f, g, MMP-2 (f) and TIMP-1 (g) concentration in the supernatant (ELISA) of cardiac fibroblasts (n = 4 biologically independent samples) without stimulus (−), with TGFβ1 or IL-11 (5 ng ml−1, 24 h). h, Il-11-neutralizing antibodies (anti-IL-11, 2 μg ml−1) block the increase in MMP-2 and TIMP-1 protein. i, In vitro monolayer scratch wound assay of cardiac fibroblasts. Wound closure was compared between stimulated (TGFβ1 or IL-11; 5 ng ml−1, 24 h) and unstimulated cardiac fibroblasts (n = 5 biologically independent samples) after 24 h. j, Cardiac fibroblasts (n = 3 biologically independent samples) were seeded in collagen gel and the contraction was monitored. The area of contraction is compared between stimulated (TGFβ1 or IL-11; 5 ng ml−1) and unstimulated groups after 72 h. k, Trans-well migration assay. After 24 h of stimulation (TGFβ1 or IL-11; 5 ng ml−1), cardiac fibroblasts (n = 6 biologically independent samples) that crossed the membrane towards either a TGFβ1- or IL-11-containing compartment were colourimetrically quantified and compared to data from unstimulated cells. l–n, Cardiac fibroblasts were incubated with TGFβ1 (5 ng ml−1, 24 h) and indicated amounts of IL11RA:gp130 decoy receptors (l; 33 amino acid (aa) or 50 aa linker peptide), anti-IL11RA antibody (m; 2 μg ml−1) or siRNA pools against IL-11 or IL11RA (n). l–n, Fibroblast activation was monitored via immunostaining for ACTA2 on the Operetta platform. decoy receptors (l): n = 7 measurements per n = 2 independent experiments; anti-IL11RA (m): n = 7 measurements per n = 2 independent experiments; siRNA (n): Operetta assay n = 7 measurements per n = 10 independent experiments. o, Human renal fibroblasts were incubated with TGFβ1 or IL-11 (5 ng ml−1, 24 h) in the presence or absence of anti-IL-11 or an IgG control antibodies (2 μg ml−1 each) for 24 h. ECM was assessed using the Operetta platform by staining for collagen I. Fluorescence was normalized to non-stimulated cells (black). p, These results were confirmed with Sirius red assay of the total collagen in the supernatant. q, rmIl-11 stimulation (5 ng ml−1, 24 h) also activated mouse cardiac and renal fibroblasts. Myofibroblasts and ECM were assessed using the Operetta platform by staining for ACTA2, collagen I or POSTN. Fluorescence was normalized to non-stimulated cells (black). o–q, These experiments were repeated three times with similar results. r, Cardiac fibroblasts analysed on the Operetta high-content imaging platform with immunostaining of ACTA2 after 24 h incubation without stimulus, TGFβ1 (5 ng ml−1, 24h) or TGFβ1 and IL-6-neutralizing antibody (2 μg ml−1, 24h). Automated quantification of fluorescence (Operetta assay n = 7 measurements per n = 6 independent experiments) shows no significant decrease in fibroblast activation using anti-IL-6 antibodies. Data are mean and circles show individual values (e) or mean ± s.d. and circles show individual values (c, d bottom right, f–h, k); box-and-whisker plots (c, d, l–n, r) show median (middle line), 25th–75th percentiles (box) and 10th–90th percentiles (whiskers). Two-tailed Dunnett’s test (c, f, g, i–k), two-tailed Student’s t-test (d, h, r) or two-tailed, Sidak-corrected Student’s t-test (l–n). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Extended Data Figure 5 IL-11 drives fibrogenic protein expression via non-canonical ERK signalling.
a, Genome-wide RNA expression differences of cardiac fibroblasts in response to IL-11 (n = 4 biologically independent samples, 5 ng ml−1, 24 h). Red indicates differentially expressed genes according to DEseq2. Fibrosis gene RNA is not increased by IL-11 treatment. b–e, RT–qPCR experiments for RNA expression of ACTA2 (b), POSTN (c), MMP2 (d) and TIMP1 (e) in response to IL-11 treatment (5 ng ml−1, 24 h) compared to unstimulated cells. IL-11 does not significantly upregulate these genes at the RNA level in cardiac fibroblasts (n = 4 biologically independent samples). f, Sirius red assay reveals significant increase in collagen protein. g, ELISA reveals increase in MMP-2 protein in the supernatant of the samples (shown in Fig. 2b, n = 6 biologically independent samples) that lack a change in RNA transcripts. h, Concentration of IL11RA (ELISA) in the supernatant of cardiac fibroblasts (n = 3 biologically independent samples) after TGFβ1 stimulation (5 ng ml−1, 24 h). i, Cardiac fibroblasts were incubated with increasing concentrations of a fusion protein consisting of IL-11 and IL11RA connected with a linker peptide that recapitulates the features of the IL-11 trans-signalling complex. Concentrations as low as 200 pg ml−1 significantly activated cardiac fibroblasts as measured using a high-content imaging platform and staining for ACTA2 expression (Operetta assay n = 7 measurements per n = 4 independent experiments). j, k, rhIL-11 (j) and hyperIL-11 (k) were added at indicated concentrations and subsequently measured using a commercially available IL-11 ELISA (n = 1 independent experiment). The ELISA did not detect rhIL-11 or hyperIL-11. We note that the reactivity to rhIL-11 was variable dependent on batch and provider and rhIL-11 was sometimes detected. However, in all experiments presented in the main figures, we confirmed that the rhIL-11 used was not detectable by the ELISA by additional measurements. This ELISA reliably detected native IL-11 secreted by human fibroblasts. l, Cardiac fibroblasts (n = 3 biologically independent samples) were incubated (8 h) with hyperIL-11 (0.2 ng ml−1) in the presence of absence of the inhibitor of protein translation, cyclohexamide (CHX, 5 μg ml−1), or protein secretion, brefeldin A (BFA, 1 μg ml−1). Both inhibitors block the increase in IL-11 protein in the supernatant in response to hyperIL-11 treatment. m, Western blot and ELISA of IL-11 in cardiac fibroblasts after hyperIL-11, the inhibitor of the Golgi secretory pathway BFA and/or the translation inhibitor CHX treatment shows de novo protein synthesis and canonical secretion of IL-11 after stimulation. n, ELISA (n = 9 biologically independent samples) and RT–qPCR (n = 5 biologically independent samples) assays show an increase in endogenous IL-11 protein but not RNA over time after rhIL-11 (5 ng ml−1) treatment. o, ERK signalling pathway activation by TGFβ1 and IL-11. Western blots show activation of the non-canonical MEK–ERK–RSK cascade in response to IL-11 stimulation of human cardiac fibroblasts. Here the response was greatest at 15 min in the two patients analysed, but more prolonged ERK activation was also seen in additional experiments. p, Downstream substrates of ERK, such as eIF4E, were also phosphorylated by rhIL-11. q, TGFβ1 also activates the ERK pathway. The time course and degree of activation was variable between patients. r, Collagen secretion (Sirius red) from cardiac fibroblasts (control, n = 6; TGFβ1, n = 6; TGFβ1 +U0126, n = 3; TGFβ1 +PD98059, n = 3; IL-11, n = 6; IL-11 +U0126, n = 3; IL-11 +PD98059, n = 3 biologically independent samples) induced by TGFβ1 or IL-11 (5 ng ml−1, 24 h) is reduced by two separate MEK inhibitors (10 μM). s, Western blot of total protein levels of key signalling molecules in fibroblasts after 24 h stimulation with AngII (100 nM), CTGF (50 ng ml−1), EDN1 (250 ng ml−1), bFGF (10 ng ml−1), IL-13 (100 ng ml−1), OSM (100 ng ml−1), PDGF (200 ng ml−1) and TGFβ1 (5 ng ml−1). The corresponding activated protein levels are shown in Fig. 2e. t, siRNA treatment of TGFβ1-stimulated cardiac fibroblasts. RT–qPCR shows SMAD-dependent upregulation of IL11 RNA (control, n = 8; TGFβ1, n = 5; siTGFB1R, n = 5; siSMAD2, n = 4; siSMAD3, n = 4 biologically independent samples). Data are mean ± s.d. (b–h, l, n, r, t); box-and-whisker plots (i) show median (middle line), 25th–75th percentiles (box) and 10th–90th percentiles (whiskers). Two-tailed Student’s t-test (b–h), two-tailed Dunnett’s test (i, r) or Sidak-corrected, two-tailed Student’s t-test (l, t) or one-way ANOVA (n). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Extended Data Figure 6 IL-11 is required for the pro-fibrotic effects of multiple stimuli.
a–h, Cardiac fibroblasts were incubated for 24 h with TGFβ1 (a; 5 ng ml−1), CTGF (b; 50 ng ml−1), PDGF (c; 200 ng ml−1), IL-13 (d; 100 ng ml−1), AngII (e; 100 nM), OSM (f; 100 ng ml−1), EDN1 (g; 250 ng ml−1) or bFGF (h; 10 ng ml−1) in the presence or absence of an IL-11-neutralizing antibody (IL-11ab) or IgG control (2 μg ml−1). Cells were stained for ACTA2, collagen I and POSTN to monitor the amount of myofibroblasts and ECM production. High-content imaging and quantification of fluorescence (Operetta assay n = 7 measurements per n = 6 independent experiments for each condition and cellular phenotype) revealed that anti-IL-11 antibodies significantly reduce the pro-fibrotic effect of these stimuli on myofibroblast ratio and ECM production. Two-tailed Dunnett’s test. Box-and-whisker plots show median (middle line), 25th–75th percentiles (box) and 10th–90th percentiles (whiskers).
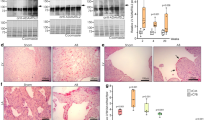
Extended Data Figure 7 Il-11 acts post-transcriptionally and causes fibrosis in vivo.
a, RNA-seq fold change in TGFβ1-regulated genes in Il11ra1+/+ cardiac fibroblasts (n = 3 biologically independent samples) compared to Il11ra1−/− cardiac fibroblasts (n = 3 biologically independent samples) after TGFβ1 stimulation (5 ng ml−1, 24 h) are highly correlated. Spearman’s correlation shows that RNA levels of fibrosis genes are upregulated equally in both genotypes. b, Wild-type and knockout fibroblasts (n = 3 biologically independent samples) were incubated with TGFβ1 (5 ng ml−1, 24 h) and RNA-seq was performed to detect differentially expressed genes using DEseq2. All genes regulated by TGFβ1 in wild-type cells are plotted with decreasing −log2(Padjusted). The P value of the same genes in stimulated Il11ra1−/− cells are plotted to the right. A similar P-value distribution suggests that TGFβ1-driven RNA expression changes are still present in the absence of IL-11 signalling showing that loss of Il11ra1 does not influence the TGFβ1-driven transcriptional response. c, d, Primary atrial fibroblasts were prepared from Il11ra1+/+ (c) or Il11ra1−/− (d) mice were incubated for 24 h without stimulus or with TGFβ1 (5 ng ml−1), IL-11 (5 ng ml−1) or AngII (100 nM). Cells were stained with antibodies against ACTA2, collagen I or POSTN. Images were taken at low magnification (10×) on the Operetta imaging platform. As shown, fibroblasts from knockout mice do not respond to pro-fibrotic stimuli at the level of pro-fibrotic protein expression. This experiment was repeated four times with similar results. e, Circulating markers of inflammation after rmIl-11 injection (100 μg kg−1 per day, three weeks; n = 14 biologically independent samples). f, Circulating levels of Tgfβ1 (ELISA) after rmIl-11 injection (control, n = 8; Il-11 injection, n = 12 biologically independent samples). g, Collagen content (HPA assay) in atrium (control, n = 7; rmIl-11, n = 10 biologically independent samples) after rmIL-11 treatment. h, The area indicative for collagen deposition was assessed over several fields in n = 4 biologically independent samples and compared between samples from rmIl-11-treated and control mice. i, Representative histological images of the heart and kidney after rmIl-11 injection indicate increased collagen content according to Masson’s trichrome staining. This experiment was repeated three times with similar results. j, RNA expression (RT–qPCR) of fibrosis genes in heart (n = 12 biologically independent samples) and kidney (n = 11 biologically independent samples) after rmIl-11 treatment compared to control. k, RNA expression (RT–qPCR) of fibrosis genes in heart (control, n = 6; Il-11-Tg, n = 3 biologically independent samples) and kidney (control, n = 7; Il-11-Tg, n = 4 biologically independent samples) of tamoxifen-treated Il-11-Tg, Col1a2–CreER and control mice. l, Cardiac fibroblasts were incubated with TGFβ1 or IL-11 (5 ng ml−1) and EdU (10 μM ml−1, 24 h), which was used to detect replicating DNA by fluorescence by automated quantification of images (Operetta assay n = 7 measurements per n = 6 independent experiments). This analysis reveals a significant increase in fibroblast proliferation (EdU+ ells) induced by both TGFβ1 and IL-11. The percentage of EdU+ cells was normalized to the average detected in non-stimulated cells. m, Cells were incubated with TGFβ1 (5 ng ml−1, 24 h) and either an IgG control or anti-IL-11 antibody (2 μg ml−1, 24 h). High-content imaging (Operetta assay n = 7 measurements per n = 6 independent experiments) and quantification of proliferating cells show that anti-IL-11 antibodies significantly reduce the effects of TGFβ1 on fibroblast proliferation. The percentage of EdU+ ells was normalized to the average detected in cells stimulated with TGFβ1 and IgG control. n, Western blots show an increase in Il-11 protein expression in the heart and kidney after tamoxifen treatment in Il-11-Tg, Col1a2–CreER mice. o, Il-11 transgenic mice were crossed with a Col1a2-promoter, tamoxifen-inducible Cre mouse strain (Il-11-Tg). Six-week-old mice were treated with tamoxifen (1 mg per day, 10 consecutive days) to induce Cre-mediated recombination. Likewise, wild-type littermates were injected with tamoxifen for 10 consecutive days as controls. The mice (control creatinine, n = 5; Il-11-Tg creatinine, n = 4; control urea, n = 6; Il-11-Tg urea, n = 4; control Tgfβ1, n = 6; Il-11-Tg Tgfβ1, n = 4 biologically independent samples) were euthanized 14 days after cessation of tamoxifen administration. Serum urea and creatinine increased and indicated renal impairment. We also observed an increase in circulating Tgfβ1 levels. p, Collagen content (HPA assay) in atrium (control, n = 11; Il-11-Tg, n = 4 biologically independent samples) from tamoxifen-treated or control Il-11-Tg mice. Data are mean ± s.d. (e, f, h, k, o); box-and-whisker plots (g, j, l, m, p) show median (middle line), 25th–75th percentiles (box) and 10th–90th percentiles (whiskers). Sidak-corrected, two-tailed Student’s t-test (e, j, k) or two-tailed Student’s t-test (f–h, m, o, p) or Dunnett’s test (l). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Extended Data Figure 8 Il-11 inhibition does not alter blood pressure after AngII treatment.
Il11ra1+/+ wild-type (n = 4 biologically independent samples) and Il11ra1−/− knockout (n = 5 biologically independent samples) mice were injected with AngII (2 mg kg−1 per day, 28 days). a, b, Systolic (a) and diastolic (b) blood pressure was measured by in vivo telemetry for one week before and four weeks after the AngII infusion. Systolic (c) and diastolic (d) blood pressure for individual mice. AngII resulted in an increase in blood pressure as expected. The genotype did not have a significant effect on blood pressure. e, Aortic root or arch velocity of the blood. There was no significant difference in the degree of aortic constriction in the TAC model between genotypes (n = 6 biologically independent samples). Two-sided Mann–Whitney U-test (e). n.s., not significant. Data are mean ± s.d. (a, b, e).
Extended Data Figure 9 Reduction in collagen deposition in Il11ra1−/− animals is independent of p38 MAPK signalling.
Il11ra1+/+ mice (control, n = 13; AngII, n = 10 biologically independent samples) and Il11ra1−/− mice (control, n = 5; AngII, n = 7 biologically independent samples) were injected with AngII (100 μg kg−1 per day, three weeks). a, HPA assay of ventricular tissue shows a decrease in collagen deposition in Il11ra1−/− mice after AngII infusion. b, Indexed heart weight of wild-type (control, n = 17; AngII, n = 17 biologically independent samples) and Il11ra1−/− (control, n = 8; AngII, n = 9 biologically independent samples) mice after AngII injection. c, Indexed heart weight of Il11ra1+/+ (control, n = 4; TAC, n = 6 biologically independent samples) and Il11ra1−/− (control, n = 6; TAC, n = 6 biologically independent samples) mice after TAC. d, Kidney weight of Il11ra1+/+ (control, n = 5; folate, n = 8 biologically independent samples) and Il11ra1−/− (control, n = 6; folate, n = 5 biologically independent samples) mice three weeks after folate injection (180 mg kg−1). a–d, Sidak-corrected, two-tailed Student’s t-test. Data are mean ± s.d. e–g, Western blot of p38 MAPK signalling in tissues of Il11ra1+/+ and Il11ra1−/− mice after AngII infusion (e), TAC (f) or folate treatment (g). h, Schematic showing the proposed role of IL-11 fibroblasts. An autocrine loop of IL-11 signalling is required to feed-forward changes in pro-fibrotic mRNA abundances to the protein level by activating translational processes that are ERK-dependent. Blocking this loop limits fibrosis caused by multiple upstream stimuli and fibrosis in preclinical models of heart and kidney disease.
Supplementary information
Supplementary Figure
This file contains source data for all western blot experiments. (PDF 575 kb)
Supplementary Table 1
Detailed information about the quality of each RNA sample, RNA-seq library and sample information about each individual that has contributed primary cells for the therapeutic target discovery high-content imaging screening and transcriptome profiling. (XLSX 71 kb)
Supplementary Table 2
Therapeutic Target Screen results: 1) Differentially expressed genes between TGFB stimulated fibroblasts and non-stimulated fibroblasts, 2) Spearman correlation (SPcor) between delta of fibroblasts expression (stimulated/non-stimulated) and delta of SMA, 3) Jensen–Shannon divergence (JSD) between of each gene across all GTEx tissues and FANTOM primary cell types (see more details in methods), 4) Average expression levels (transcripts per million, TPM) in TGFB1 stimulated and non-stimulated (baseline only) fibroblasts. Log2 fold change, shrunken Log2-fold changes computed by DESeq2 package. BH adj.P, Benjamini-Hochberg (BH) adjusted p-value. (XLSX 1759 kb)
Supplementary Table 3
Gene Ontology database gene set enrichment analysis (GSEA) results for the stimulated versus baseline fibroblasts (GSEA computed by ranking all the genes by DESeq output statistic). Only terms enriched with FDR < 0.05 are presented. NES denotes normalized enrichment score. (XLSX 191 kb)
Source data
Rights and permissions
About this article
Cite this article
Schafer, S., Viswanathan, S., Widjaja, A. et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature 552, 110–115 (2017). https://doi.org/10.1038/nature24676
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24676
This article is cited by
-
Fibroblast-specific PRMT5 deficiency suppresses cardiac fibrosis and left ventricular dysfunction in male mice
Nature Communications (2024)
-
ALKBH5-mediated m6A modification of IL-11 drives macrophage-to-myofibroblast transition and pathological cardiac fibrosis in mice
Nature Communications (2024)
-
A Multimodal Omics Framework to Empower Target Discovery for Cardiovascular Regeneration
Cardiovascular Drugs and Therapy (2024)
-
IRF4 Participates in Pulmonary Fibrosis Induced by Silica Particles through Regulating Macrophage Polarization and Fibroblast Activation
Inflammation (2024)
-
Emerging therapeutic targets in systemic sclerosis
Journal of Molecular Medicine (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.