Abstract
Maternally inherited 15q11-13 chromosomal triplications cause a frequent and highly penetrant type of autism linked to increased gene dosages of UBE3A, which encodes a ubiquitin ligase with transcriptional co-regulatory functions. Here, using in vivo mouse genetics, we show that increasing UBE3A in the nucleus downregulates the glutamatergic synapse organizer Cbln1, which is needed for sociability in mice. Epileptic seizures also repress Cbln1 and are found to expose sociability impairments in mice with asymptomatic increases in UBE3A. This Ube3a–seizure synergy maps to glutamate neurons of the midbrain ventral tegmental area (VTA), where Cbln1 deletions impair sociability and weaken glutamatergic transmission. We provide preclinical evidence that viral-vector-based chemogenetic activation of, or restoration of Cbln1 in, VTA glutamatergic neurons reverses the sociability deficits induced by Ube3a and/or seizures. Our results suggest that gene and seizure interactions in VTA glutamatergic neurons impair sociability by downregulating Cbln1, a key node in the expanding protein interaction network of autism genes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chen, J. A., Peñagarikano, O., Belgard, T. G., Swarup, V. & Geschwind, D. H. The emerging picture of autism spectrum disorder: genetics and pathology. Annu. Rev. Pathol. 10, 111–144 (2015)
El Achkar, C. M. & Spence, S. J. Clinical characteristics of children and young adults with co-occurring autism spectrum disorder and epilepsy. Epilepsy Behav. 47, 183–190 (2015)
Glessner, J. T. et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459, 569–573 (2009)
Smith, S. E. et al. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci. Transl. Med. 3, 103ra97 (2011)
Jiang, Y. H. et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 21, 799–811 (1998)
Kaphzan, H. et al. Genetic reduction of the α1 subunit of Na/K-ATPase corrects multiple hippocampal phenotypes in Angelman syndrome. Cell Reports 4, 405–412 (2013)
Matsuura, T. et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat. Genet. 15, 74–77 (1997)
Wallace, M. L., Burette, A. C., Weinberg, R. J. & Philpot, B. D. Maternal loss of Ube3a produces an excitatory/inhibitory imbalance through neuron type-specific synaptic defects. Neuron 74, 793–800 (2012)
Scheffner, M., Huibregtse, J. M., Vierstra, R. D. & Howley, P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75, 495–505 (1993)
Nawaz, Z. et al. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol. Cell. Biol. 19, 1182–1189 (1999)
Scoles, H. A., Urraca, N., Chadwick, S. W., Reiter, L. T. & Lasalle, J. M. Increased copy number for methylated maternal 15q duplications leads to changes in gene and protein expression in human cortical samples. Mol. Autism 2, 19 (2011)
Basu, S. N., Kollu, R. & Banerjee-Basu, S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 37, D832–D836 (2009)
Matsuda, K. & Yuzaki, M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur. J. Neurosci. 33, 1447–1461 (2011)
Wei, P. et al. The Cbln family of proteins interact with multiple signaling pathways. J. Neurochem. 121, 717–729 (2012)
Elegheert, J. et al. Structural basis for integration of GluD receptors within synaptic organizer complexes. Science 353, 295–299 (2016)
Matsuda, K. et al. Cbln1 is a ligand for an orphan glutamate receptor δ2, a bidirectional synapse organizer. Science 328, 363–368 (2010)
Uemura, T. et al. Trans-synaptic interaction of GluRδ2 and neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 141, 1068–1079 (2010)
Ryu, K., Yokoyama, M., Yamashita, M. & Hirano, T. Induction of excitatory and inhibitory presynaptic differentiation by GluD1. Biochem. Biophys. Res. Commun. 417, 157–161 (2012)
Hirai, H. et al. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat. Neurosci. 8, 1534–1541 (2005)
Bao, D. et al. Cbln1 is essential for interaction-dependent secretion of Cbln3. Mol. Cell. Biol. 26, 9327–9337 (2006)
Grier, M. D., Carson, R. P. & Lagrange, A. H. Of mothers and myelin: Aberrant myelination phenotypes in mouse model of Angelman syndrome are dependent on maternal and dietary influences. Behav. Brain Res. 291, 260–267 (2015)
Yang, M., Silverman, J. L. & Crawley, J. N. Automated three-chambered social approach task for mice. Curr. Protoc. Neurosci. Chapter 8, Unit 8.26 (2011)
Yadav, R. et al. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS One 7, e32969 (2012)
Iijima, T., Emi, K. & Yuzaki, M. Activity-dependent repression of Cbln1 expression: mechanism for developmental and homeostatic regulation of synapses in the cerebellum. J. Neurosci. 29, 5425–5434 (2009)
Takechi, K., Suemaru, K., Kiyoi, T., Tanaka, A. & Araki, H. The α4β2 nicotinic acetylcholine receptor modulates autism-like behavioral and motor abnormalities in pentylenetetrazol-kindled mice. Eur. J. Pharmacol. 775, 57–66 (2016)
Taylor, S. R. et al. GABAergic and glutamatergic efferents of the mouse ventral tegmental area. J. Comp. Neurol. 522, 3308–3334 (2014)
Zhang, S. et al. Dopaminergic and glutamatergic microdomains in a subset of rodent mesoaccumbens axons. Nat. Neurosci. 18, 386–392 (2015)
Gunaydin, L. A. et al. Natural neural projection dynamics underlying social behavior. Cell 157, 1535–1551 (2014)
Yamaguchi, T., Qi, J., Wang, H. L., Zhang, S. & Morales, M. Glutamatergic and dopaminergic neurons in the mouse ventral tegmental area. Eur. J. Neurosci. 41, 760–772 (2015)
Greer, P. L. et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating Arc. Cell 140, 704–716 (2010)
Khan, O. Y. et al. Multifunction steroid receptor coactivator, E6-associated protein, is involved in development of the prostate gland. Mol. Endocrinol. 20, 544–559 (2006)
Kühnle, S ., Mothes, B ., Matentzoglu, K & Scheffner, M. Role of the ubiquitin ligase E6AP/UBE3A in controlling levels of the synaptic protein Arc. Proc. Natl Acad. Sci. USA 110, 8888–8893 (2013)
Kim, S., Chahrour, M., Ben-Shachar, S. & Lim, J. Ube3a/E6AP is involved in a subset of MeCP2 functions. Biochem. Biophys. Res. Commun. 437, 67–73 (2013)
Lyst, M. J. et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat. Neurosci. 16, 898–902 (2013)
McClelland, S. et al. The transcription factor NRSF contributes to epileptogenesis by selective repression of a subset of target genes. eLife 3, e01267 (2014)
Liu, M. et al. Neuronal conditional knockout of NRSF decreases vulnerability to seizures induced by pentylenetetrazol in mice. Acta Biochim. Biophys. Sin. (Shanghai) 44, 476–482 (2012)
Kent, W. J. et al. The human genome browser at UCSC. Genome Res/ 12, 996–1006 (2002)
Yi, J. J. et al. An autism-linked mutation disables phosphorylation control of UBE3A. Cell 162, 795–807 (2015)
Ronchi, V. P., Klein, J. M., Edwards, D. J. & Haas, A. L. The active form of E6-associated protein (E6AP)/UBE3A ubiquitin ligase is an oligomer. J. Biol. Chem. 289, 1033–1048 (2014)
Gossan, N. C. et al. The E3 ubiquitin ligase UBE3A is an integral component of the molecular circadian clock through regulating the BMAL1 transcription factor. Nucleic Acids Res. 42, 5765–5775 (2014)
Berrios, J. et al. Loss of UBE3A from TH-expressing neurons suppresses GABA co-release and enhances VTA-NAc optical self-stimulation. Nat. Commun. 7, 10702 (2016)
Riday, T. T. et al. Pathway-specific dopaminergic deficits in a mouse model of Angelman syndrome. J. Clin. Invest. 122, 4544–4554 (2012)
Zhou, Y. D. et al. Arrested maturation of excitatory synapses in autosomal dominant lateral temporal lobe epilepsy. Nat. Med. 15, 1208–1214 (2009)
Anderson, M. P . et al. Thalamic Cav3.1 T-type Ca2+ channel plays a crucial role in stabilizing sleep. Proc. Natl Acad. Sci. USA 102, 1743–1748 (2005)
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001)
Salvat, C., Wang, G., Dastur, A., Lyon, N. & Huibregtse, J. M. The -4 phenylalanine is required for substrate ubiquitination catalyzed by HECT ubiquitin ligases. J. Biol. Chem. 279, 18935–18943 (2004)
Vong, L. et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71, 142–154 (2011)
Tong, Q. et al. Synaptic glutamate release by ventromedial hypothalamic neurons is part of the neurocircuitry that prevents hypoglycemia. Cell Metab. 5, 383–393 (2007)
Gorski, J. A. et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314 (2002)
Rose, M. F ., Ahmad, K. A ., Thaller, C & Zoghbi, H. Y. Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proc. Natl Acad. Sci. USA 106, 22462–22467 (2009)
Zhuang, X., Masson, J., Gingrich, J. A., Rayport, S. & Hen, R. Targeted gene expression in dopamine and serotonin neurons of the mouse brain. J. Neurosci. Methods 143, 27–32 (2005)
Kobayashi, K. et al. Survival of developing motor neurons mediated by Rho GTPase signaling pathway through Rho-kinase. J. Neurosci. 24, 3480–3488 (2004)
Rossi, J. et al. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 13, 195–204 (2011)
Scott, M. M., Williams, K. W., Rossi, J., Lee, C. E. & Elmquist, J. K. Leptin receptor expression in hindbrain Glp-1 neurons regulates food intake and energy balance in mice. J. Clin. Invest. 121, 2413–2421 (2011)
Madisen, L. et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci. 15, 793–802 (2012)
Rozen, S & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 (2000)
Ye, J. et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134 (2012)
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protocols 3, 1101–1108 (2008)
The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000)
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004)
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L. & Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011)
Silverman, J. L. et al. GABAB receptor agonist R-baclofen reverses social deficits and reduces repetitive behavior in two mouse models of autism. Neuropsychopharmacology 40, 2228–2239 (2015)
Allensworth, M., Saha, A., Reiter, L. T. & Heck, D. H. Normal social seeking behavior, hypoactivity and reduced exploratory range in a mouse model of Angelman syndrome. BMC Genet. 12, 7 (2011)
Yang, M. & Crawley, J. N. Simple behavioral assessment of mouse olfaction. Curr. Protoc. Neurosci. Chapter 8, Unit 8.24 (2009)
Ferraro, T. N . et al. Mapping loci for pentylenetetrazol-induced seizure susceptibility in mice. J. Neurosci. 19, 6733–6739 (1999)
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007)
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012)
Acknowledgements
We thank O. DiStefano, G. Salimando and R. Broadhurst for colony work and the Harvard Medical School Neurobiology Imaging Facility (NINDS P30 Core Center Grant NS07203) and Boston Children’s Hospital IDDRC (1U54HD090255, P30HD18655). Supported by funding to V.K. (1R25NS070682 and an American Academy of Neurology Research Training Fellowship) and to M.P.A. from the NIH (1R01NS08916, 1R21MH100868, 1R21HD079249), Nancy Lurie Marks Family Foundation, Landreth Foundation, Simons Foundation, and Autism Speaks/National Alliance for Autism Research. R.A. was supported by Klarman Family Foundation.
Author information
Authors and Affiliations
Contributions
V.K., D.C.S., Y.N. and M.P.A. designed the study. V.K. and M.P.A wrote the manuscript. V.K., D.C.S. and Y.N. performed all experiments and analyses except for the following: E.O., M.J.S.N. and T.J.C validated microarray-identified gene regulations by qRT–PCR; F.M., R.A. and M.P.A. performed Gene Ontology and cluster visualization; I.N. performed nuclear/cytosolic fractionation studies; S.P. performed olfaction and open-field experiments; M.A.J., B.L.T., M.A.S., E.M.K. and M.J.S.N. developed molecular probes and constructs.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks E. Kim and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Protein–protein interactions between genes regulated in Ube3a-2× mice and autism-related genes.
Edges between gene-labelled nodes represent documented protein–protein physical interactions. Clusters shown here are labelled with their major statistically significant Gene Ontology classification(s) together with P values representing significant enrichment. Within each cluster, all the genes (UBE3A-regulated and autism-related) were analysed.
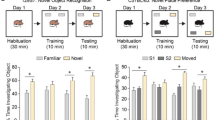
Extended Data Figure 2 Ube3a-NLS mice and mice with increased untagged UBE3A levels display impaired sociability.
a, Total UBE3A protein levels by western blot in wild-type, Ube3a-1×, Ube3a-NLS3 and Ube3a-NLS7 mice (F[3,17] = 12.30, P < 0.001, 5 females per group). b, Flag staining in Ube3a-1× and Ube3a-NLS mice, counterstained with DAPI (somatosensory cortex; 3 females per genotype). Scale bar, 50 μm. c, Quantified nuclear and neuropil Flag immunofluorescence in arbitrary units (a.u.). d, UBE3A protein levels in cytosolic and nuclear fractions obtained from wild-type and Ube3a-NLS7 mice (4 females per group), demonstrating a selective increase in nuclear UBE3A protein levels (see Supplementary Fig. 1). TBP, TATA-box binding protein. e, Chamber times (for Fig. 1d) during the social trial showing absolute times spent in social, middle and opposite chambers. f, Distances moved during acclimation/social trials analysed by two-way ANOVA: WT/Ube3a-1× (trial factor F[1,56] = 18.09, P < 0.001, genotype factor F[1,56] = 0.73, P > 0.1), WT/Ube3a-2× (trial factor F[1,52] = 12.2, P < 0.001, genotype factor F[1,52] = 2.046, P > 0.1), WT/Ube3a-2× untagged (trial factor F[1,50] = 12.21, P < 0.01, genotype factor F[1,50] = 9.67, P < 0.01), WT/Ube3a-NLS3 (trial factor F[1,56] = 14.41, P < 0.001), genotype factor F[1,56] = 0.53, P > 0.1), WT/Ube3a-NLS7 (trial factor F[1,56] = 22.68, P < 0.001, genotype factor F[1,56] = 4.88, P < 0.05), WT/p-Ube3a-mKO (trial factor F[1,62] = 3.67, P > 0.05, genotype factor F[1,62] = 216.7, P < 0.0001), WT/m-Ube3a-mKO (trial factor F[1,48] = 1.75, P > 0.1, genotype factor F[1,48] = 197.9, P < 0.001). g, Total time spent physically interacting during genotype-matched paired female interactions (for Fig. 1e). h, Time spent self-grooming by female mice during a 10-min observation session across Ube3a-1× (n = 10 versus n = 14 WT), Ube3a-2× (n = 11 versus n = 12 WT), Ube3a-NLS3 (n = 14 per group) and Ube3a-NLS7 (n = 14 per group) mice. i, Ube3a-NLS7 (n = 15) and wild-type (n = 12) littermates take similar times to find a food item buried in fresh bedding. j, k, In an open field, Ube3a-NLS7 and wild-type (n = 5 per group) littermates spend equal times in the centre of an open field (j) and ambulate equal distances (k). l, m, In an elevated plus maze test, Ube3a-NLS7 mice (n = 10) spend significantly less time in the open arms (l) and ambulate slightly shorter distances overall than wild-type littermates (n = 7) (m). Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. Two-tailed unpaired Student’s t-tests are used for all comparisons unless otherwise specified.
Extended Data Figure 3 qPCR-based validation of various genes downregulated in cortical samples from Ube3a-2× mice.
a, b, In separate cohorts of wild-type and transgenic or maternal knockout Ube3a mice, we confirmed the downregulation of the following genes: Fat2, Arhgef33, Crtam, Dao, Smpx, Pcp2, Gsbs (also known as Ppp1r17) (a), A2m, Mab21l1, Zic1, Car8, Lhx1, Calb2, Slc1a6, Chn2 and St18 (b). Sample sizes: Ube3a-1× (ref. 4), Ube3a-NLS, Ube3a-2× (ref. 4) and m-Ube3a-mKO (ref. 21) (n = 3 pooled samples from two mice each); Ube3a-2× (untagged) and p-Ube3a-mKO (ref. 5) (n = 6–12 per group, unpooled). Data are mean ± s.e.m. Transgene constructs and/or breeding schemes are shown on the right. *P < 0.05, **P < 0.01, ***P < 0.001. Unpaired two-tailed Students t-tests.
Extended Data Figure 4 qRT–PCR-based validation of various genes upregulated in cortical samples from Ube3a-2× mice.
a, In separate cohorts of wild-type and Ube3a-2× mice, the upregulation of the following genes was confirmed by qPCR: S100a8, S100a9, Gh, Cdhr1, Nptx2, Blnk, Islr2, Grasp, Agmat, Pcdh8, Rasl10a, Dlx1, Wisp1, Glt8d2, Igfbp4, Ptgs2, Dmkn, Gcnt2, Pcdh15, Efna3, Kalrn, Tnnt2, Kcnh5, Prkg2, Trpc6, Kcnq3 and Homer2. b–d, Regulation of these genes in cortical samples from Ube3a-NLS7 (b), Ube3a-1× (c) and Ube3a-mKO (d) mice. n = 3 per genotype, with each biological replicate representing a pooled sample of tissue from two genotype-matched mice. Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. Unpaired two-tailed Students t-tests.
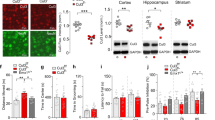
Extended Data Figure 5 Characterization of VGLUT2-cre;Cbln1fl/fl mice.
a, Representative coronal sections from the Allen Mouse Brain Atlas66, with shaded red regions depicting the approximate geometric centre of punch biopsy samples used for qPCR experiments. Image credit: Allen Institute for Brain Science. b, Chamber times (for Fig. 1g) during the social trial showing absolute time spent in social, middle and opposite chambers. c, Distances moved during acclimation/social trials analysed by two-way ANOVA (trial factor F[1,32] = 27.21, P < 0.0001, genotype factor F[1,32] = 8.186, P < 0.01). d, Total time spent physically interacting during genotype-matched paired female interactions (for Fig. 1h, male–female pairs: Cre− n = 14, Cre+ n = 8, genotype-matched female pairs: Cre− n = 11, Cre+ n = 16). e, Total time spent self-grooming during a 10-min observation period (males only, n = 8 per group). f, g, In the elevated plus maze, VGLUT2-cre;Cbln1fl/fl (n = 10) mice spent greater time in the open arms than Cbln1fl/fl (WT) mice (n = 8) (f) and ambulated significantly greater distances (g). h, Performance of VGLUT2-cre;Cbln1fl/fl (n = 16) and Cbln1fl/fl (n = 13) mice on buried food testing. i, j, Open-field testing of VGLUT2-cre;Cbln1fl/fl and Cbln1fl/fl mice (n = 10 per group) with time in centre (i) and distances moved (j). k, VGLUT2-cre;Cbln1fl/fl mice (n = 16) display reduced latencies to fall off of an accelerating rotarod compared with Cbln1fl/fl mice (two-way repeated measures ANOVA, n = 13 per group, genotype factor, F[1, 81] = 18.10, P < 0.001). Data are mean ± s.e.m. *P < 0.05, **P < 0.01. Two-tailed unpaired Student’s t-tests are used for all comparisons unless otherwise specified.
Extended Data Figure 6 Recurrent seizures induced by PTZ impair sociability.
a, Ten daily 30 mg kg−1 PTZ injections applied to male wild-type mice resulted in a gradual daily increase in the percentage of animals that displayed a PTZ-induced generalized convulsion (n = 25). b, Weight change for PTZ- (n = 10) or saline- (n = 8) treated mice, with test day factor analysed by two-way repeated measures ANOVA (F[9,160] = 0.28, P > 0.1, treatment factor F[1,160] = 0.34, P > 0.1). c, d, 24 h after the last PTZ (n = 16) or saline (n = 14) injection, open-field testing identified no significant differences in the time spent in the centre of the field (c) or the total distance ambulated in the field (d) (n = 10 per group). e, f, Similarly, on the elevated plus maze, 10 PTZ-induced seizures did not affect the time spent in the open arms (e) or distance moved (f) (n = 10 per group). g, Distances moved during acclimation/social trials for male mice analysed by two-way ANOVA (for Fig. 2b, treatment factor F[1,32] = 0.38, P > 0.1, trial factor F[1,32] = 0.06, P > 0.1). h, i, Female wild-type FVB mice exposed to an identical 10-day PTZ protocol (n = 10) demonstrated impaired sociability 24 h after the last injection compared with mice that received saline alone (n = 9) (h), whereas the distances moved during acclimation/social trials were not affected (treatment factor F[1,34] = 2.47, P > 0.1, trial factor F[1,34] = 1.79, P > 0.1) (i). j, k, Ten PTZ-induced seizures do not affect self-grooming rates (n = 10 females per group) (j) or performance on the buried food test (k) (males: saline n = 10, PTZ n = 7 per group, females: saline n = 7, PTZ n = 9 per group). l, m, 30 days after the last of 10 daily PTZ injections, male wild-type FVB mice continue to display impaired sociability (saline n = 10 per group, PTZ n = 7 per group) (l), as do daily PTZ-treated female mice (saline n = 6 per group, PTZ n = 7 per group) (m). n, Distances moved during acclimation/social trials for Fig. 2f, g, analysed by two-way ANOVA. WT/p-Ube3a-mKO (trial factor F[1,62] = 5.06, P < 0.05, genotype factor F[1,62] = 209.2, P < 0.0001) and WT/m-Ube3a-mKO (trial factor F[1,54] = 0.20, P > 0.1, genotype factor F[1,54] = 56.06, P < 0.001). o, Distances moved for Fig. 2h (trial factor F[1.62] = 3.57, P > 0.05, group factor F[2,62] = 1.57, P > 0.05). p, Distances moved for Fig. 2i (trial factor F[1,62] = 13.29, P < 001, group factor F[2,62] = 2.43, P > 0.05). q, Ube3a mRNA measured in punches from wild-type FVB males treated with 10 daily injections of 30 mg kg−1 PTZ or saline (HPc, n = 8 per group, ECx and VTA: saline n = 10, PTZ n = 8). Data are mean ± s.e.m. ***P < 0.001. Two-tailed unpaired Student’s t-tests are used for all comparisons unless otherwise specified.
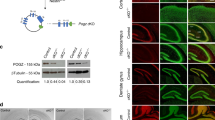
Extended Data Figure 7 Validation oflox-TB-Ube3a mice.
a, qPCR analysis of Ube3a mRNA from hippocampal samples (Extended Data Fig. 5a) demonstrating the upregulation of Ube3a in VGLUT2-cre;lox-TB-Ube3a and Emx-cre;lox-TB-Ube3a mice. b, Similar analysis of Ube3a mRNA levels from tissue punches of the VTA of DAT-cre;lox-TB-Ube3a mice (n = 8 per group). c–h, Spatial confirmation of Flag staining in various crosses of lox-TB-Ube3a mice, confirming expression of UBE3A in inhibitory forebrain neurons (c), cortical and hippocampal neurons (d), scattered staining in midbrain and pontine nuclei (e), locus coeruleus (f), striatal cholinergic interneurons (g), and raphe regions (h). Images are representative of n = 2–3 mice per group. Data are mean ± s.e.m. *P < 0.05, ***P < 0.001. Two-tailed unpaired Student’s t-tests.
Extended Data Figure 8 Targeted overexpression of Ube3a in VGLUT2+ and DAT+ neurons increases the susceptibility to seizure-induced deficits in sociability.
a–h, For each line of mice, we provide baseline sniffing time measures on sociability testing (far left), distances moved during this baseline test (second from left), seizure severity scores during five (alternate day) 30 mg kg−1 PTZ injections (middle), post-seizure sniffing time measures on three-chamber testing (second from right) and associated distances moved (far right). For sociability data, black and white bars represent sniffing times for social and opposite wire mesh cages, respectively. As described in Fig. 1, sniffing time is analysed by two-way repeated-measures ANOVA. F statistics and P values for ‘interaction effects’ between chamber side (social versus opposite) or experimental group (viruses, treatments or genotypes) and ‘main effects’ are shown in Supplementary Table 5. For significant interaction and/or main effects, a Bonferroni post-hoc test P value is reported (adjusted for multiple comparisons). For distance measures, black and white bars represent distances moved during acclimation and social trials, respectively. For VGLUT2-cre and DAT-cre lines, see Fig. 3c–e, g–i. Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Data Figure 9 Stereotaxic manipulations of Ube3a, VGLUT2 and Cbln1 in VTA.
a, qPCR analysis of Cbln1 mRNA in VTA tissue punches of wild-type and Ube3a-1× (ref. 4) mice obtained 24 h after the last of 5 alternate day PTZ injections. b, Distances moved during acclimation/social trials for mice in Fig. 3k (genotype factor F[1,28] = 14.45, P < 0.001, trial factor F[1,28] = 5.16, P < 0.05). c, Distances moved for Fig. 3l (genotype factor F[1,28] = 1.267, P > 0.1, trial factor F[1,28] = 2.72, P > 0.1). d–f, Distances moved for Fig. 4a (virus factor F[1,34] = 2.81, P > 0.1, trial factor F[1,34] = 2.02, P > 0.1) (d), time spent in the centre (e), or total distance moved (f) in an open field for AAV-Cre-GFP-injected (n = 12) or AAV-GFP-injected (n = 8) Cbln1fl/fl mice. g, Distances moved for Fig. 4b (virus factor F[1,44] = 7.9, P < 0.01, trial factor F[1,44] = 20.49, P < 0.001). h, Distances moved for Fig. 4c (genotype factor F[1,26] = 0.22, P > 0.1, trial factor F[1,26] = 5.86, P < 0.05). i, Cumulative frequency plots of spontaneous EPSC (sEPSC) amplitudes for Fig. 4d, e. j, Representative response of an MSN to injected currents (−20 to +280 pA). k, Average resting membrane potentials in the NAc medial shell in control mice (n = 23 neurons) and mice with VTA-targeted AAV-Cre-GFP-mediated deletions of Cbln1 (n = 22 neurons, P > 0.1). l, Laser intensity needed to induce EPSC currents in medial shell NAc MSNs from ChR2-eYFP mice with AAV-Cre-GFP infused into the VTA (n = 5 neurons). m, Distances moved for Fig. 4i (trial factor F[1,36] = 9.45, P < 0.01, virus factor F[1,36] = 8.49, P < 0.01). n, o, Time spent in the centre (n) and distances moved (o) during the open field test after AAV-Cre-GFP-mediated deletion of VGLUT2 in the VTA (n = 8). p, Distances moved for Fig. 4i (genotype factor F[1,36] = 1.15, P > 0.1, trial factor F[1,36] = 6.18, P < 0.1). Data are mean ± s.e.m. *P < 0.05. Two-tailed unpaired Student’s t-tests are used for all comparisons across two groups, whereas two-way ANOVA is used for all comparisons of distances moved. Kolmogorov–Smirnov test was used in i.
Extended Data Figure 10 Chemogenetic manipulations of VTA VGLUT2+ neurons.
a, Distances moved for Fig. 5a (trial factor F[1,48] = 20.11, P < 0.001, treatment factor F[1,48] = 0.73, P > 0.5). b, Distances moved for Fig. 5b (treatment factor F[2,111] = 1.04, P > 0.5, trial factor F[2,111] = 9.55, P < 0.01). c, Distances moved for Fig. 5c (trial factor F[1,102] = 18.16, P < 0.0001, group factor F[2,102] = 5.7, P < 0.01). d, Distances moved for Fig. 5d (trial factor F[1,90] = 3.64, P > 0.05, group factor F[2,90] = 0.77, P > 0.05). e, Left, distances moved for Fig. 5e, left (baseline) (trial factor F[1,52] = 7.08, P < 0.05, virus factor F[1.52] = 0.022, P > 0.5). Right, distances moved for Fig. 5e, right (after PTZ) (trial factor F[1,76] = 8.48, P < 0.01, virus factor F[1,76] = 1.16, P > 0.05). f, qPCR analysis of Rest (also known as Nrsf) mRNA levels in VTA 24 h after 10 PTZ (n = 8) or saline (n = 10) injections. g, Data presented from the Encode Project Consortium67 showing locations of REST binding within Cbln1 intron of human (top) and mouse (bottom). Data are mean ± s.e.m. *P < 0.05. Two-tailed unpaired Student’s t-tests are used for all comparisons across two groups, whereas two-way ANOVA is used for all comparisons of distances moved.
Supplementary information
Supplementary Figure 1
This file shows the source gel for the representative bands provided in Extended Data Figure 2d. (PDF 140 kb)
Supplementary Table 1
This file lists genes identified as up or downregulated in cortical samples from Ube3a2x mice compared with WT littermates. A +/- 1.4 fold change cutoff was used. Accession numbers, fold change and p values are shown. (XLSX 46 kb)
Supplementary Table 2
This file lists microarray-identified genes by gene ontology classifications together with a calculated p value for enrichment within a specific biological process. Worksheet 1 within this table displays results of annotation through the Kyoto Encyclopedia of Genes and Genomes (KEGG). Worksheets 2-4 display the same results when gene lists were annotated by classifications as defined by the Gene Ontology Consortium (biological process, cellular component and molecular function). (XLSX 3250 kb)
Supplementary Table 3
This file contains a list of selected 276 genes identified as strongly linked to autism from a larger list of 706 genes from the Simons Foundation Autism Research Initiative (SFARI, https://gene.sfari.org/autdb/HG_Home.do - see Basu, S. N., Kollu, R. & Banerjee-Basu, S. AutDB: a gene reference resource for autism research. Nucleic acids research 37, D832-836, doi:10.1093/nar/gkn835 (2009)). (XLSX 34 kb)
Supplementary Table 4
This file contains sequences for forward and reverse primers utilized in quantitative real-time PCR experiments. (XLSX 37 kb)
Supplementary Table 5
This file comprises: Worksheet 1 - A table of chamber times (in seconds) provided for each graph where "sniffing times" are depicted in the main and extended figures. This excludes Fig. 1d and Fig. 1h, where chamber times are depicted graphically in Extended Data Figs. 2e and 5b respectively; Worksheet 2 - Distances moved, chamber times and “sniffing times” (for "social" and "opposite") for a population of 101 wildtype seizure naïve FVB mice (males and females) with column statistics (mean and measures of variance) as well as results of Kolmogorov-Smirnov testing for the presence of a normal distribution; Worksheet 3 - Two-way repeated measures ANOVA analysis results for each graph depicting three chamber sociability data. The results of post-hoc testing are provided comparing "social" versus "opposite" sniffing times for each group separately. (XLSX 66 kb)
Supplementary Table 6
This file contains experimental data. (XLSX 667 kb)
Source data
Rights and permissions
About this article
Cite this article
Krishnan, V., Stoppel, D., Nong, Y. et al. Autism gene Ube3a and seizures impair sociability by repressing VTA Cbln1. Nature 543, 507–512 (2017). https://doi.org/10.1038/nature21678
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21678
This article is cited by
-
mRNA nuclear retention reduces AMPAR expression and promotes autistic behavior in UBE3A-overexpressing mice
EMBO Reports (2024)
-
C1ql1-Bai3 signaling is necessary for climbing fiber synapse formation in mature Purkinje cells in coordination with neuronal activity
Molecular Brain (2023)
-
Promising therapeutic aspects in human genetic imprinting disorders
Clinical Epigenetics (2022)
-
Adolescent sleep shapes social novelty preference in mice
Nature Neuroscience (2022)
-
Shank2/3 double knockout-based screening of cortical subregions links the retrosplenial area to the loss of social memory in autism spectrum disorders
Molecular Psychiatry (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.