Abstract
Metabolic pathways that contribute to adiposity and ageing are activated by the mammalian target of rapamycin complex 1 (mTORC1) and p70 ribosomal protein S6 kinase 1 (S6K1) axis1,2,3. However, known mTORC1–S6K1 targets do not account for observed loss-of-function phenotypes, suggesting that there are additional downstream effectors of this pathway4,5,6. Here we identify glutamyl-prolyl-tRNA synthetase (EPRS) as an mTORC1–S6K1 target that contributes to adiposity and ageing. Phosphorylation of EPRS at Ser999 by mTORC1–S6K1 induces its release from the aminoacyl tRNA multisynthetase complex, which is required for execution of noncanonical functions of EPRS beyond protein synthesis7,8. To investigate the physiological function of EPRS phosphorylation, we generated Eprs knock-in mice bearing phospho-deficient Ser999-to-Ala (S999A) and phospho-mimetic (S999D) mutations. Homozygous S999A mice exhibited low body weight, reduced adipose tissue mass, and increased lifespan, similar to S6K1-deficient mice9,10,11 and mice with adipocyte-specific deficiency of raptor, an mTORC1 constituent12. Substitution of the EprsS999D allele in S6K1-deficient mice normalized body mass and adiposity, indicating that EPRS phosphorylation mediates S6K1-dependent metabolic responses. In adipocytes, insulin stimulated S6K1-dependent EPRS phosphorylation and release from the multisynthetase complex. Interaction screening revealed that phospho-EPRS binds SLC27A1 (that is, fatty acid transport protein 1, FATP1)13,14,15, inducing its translocation to the plasma membrane and long-chain fatty acid uptake. Thus, EPRS and FATP1 are terminal mTORC1–S6K1 axis effectors that are critical for metabolic phenotypes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Johnson, S. C., Rabinovitch, P. S. & Kaeberlein, M. mTOR is a key modulator of ageing and age-related disease. Nature 493, 338–345 (2013)
Lamming, D. W. & Sabatini, D. M. A central role for mTOR in lipid homeostasis. Cell Metab. 18, 465–469 (2013)
Fontana, L., Partridge, L. & Longo, V. D. Extending healthy life span—from yeast to humans. Science 328, 321–326 (2010)
Tumaneng, K., Russell, R. C. & Guan, K. L. Organ size control by Hippo and TOR pathways. Curr. Biol. 22, R368–R379 (2012)
Magnuson, B., Ekim, B. & Fingar, D. C. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 441, 1–21 (2012)
Ruvinsky, I. & Meyuhas, O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci. 31, 342–348 (2006)
Arif, A. et al. Two-site phosphorylation of EPRS coordinates multimodal regulation of noncanonical translational control activity. Mol. Cell 35, 164–180 (2009)
Arif, A., Jia, J., Moodt, R. A., DiCorleto, P. E. & Fox, P. L. Phosphorylation of glutamyl-prolyl tRNA synthetase by cyclin-dependent kinase 5 dictates transcript-selective translational control. Proc. Natl Acad. Sci. USA 108, 1415–1420 (2011)
Um, S. H. et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431, 200–205 (2004)
Selman, C. et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326, 140–144 (2009)
Carnevalli, L. S. et al. S6K1 plays a critical role in early adipocyte differentiation. Dev. Cell 18, 763–774 (2010)
Polak, P. et al. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 8, 399–410 (2008)
Wu, Q. et al. FATP1 is an insulin-sensitive fatty acid transporter involved in diet-induced obesity. Mol. Cell. Biol. 26, 3455–3467 (2006)
Stahl, A., Evans, J. G., Pattel, S., Hirsch, D. & Lodish, H. F. Insulin causes fatty acid transport protein translocation and enhanced fatty acid uptake in adipocytes. Dev. Cell 2, 477–488 (2002)
Kim, J. K. et al. Inactivation of fatty acid transport protein 1 prevents fat-induced insulin resistance in skeletal muscle. J. Clin. Invest. 113, 756–763 (2004)
Guo, M., Yang, X. L. & Schimmel, P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 11, 668–674 (2010)
Ray, P. S. & Fox, P. L. Origin and evolution of glutamyl-prolyl tRNA synthetase WHEP domains reveal evolutionary relationships within Holozoa. PLoS One 9, e98493 (2014)
Ma, X. M. & Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318 (2009)
Ge, Y. et al. mTOR regulates skeletal muscle regeneration in vivo through kinase-dependent and kinase-independent mechanisms. Am. J. Physiol. Cell Physiol. 297, C1434–C1444 (2009)
Ohanna, M. et al. Atrophy of S6K1−/− skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat. Cell Biol. 7, 286–294 (2005)
Yi, S. A. et al. S6K1 phosphorylation of H2B mediates EZH2 trimethylation of H3: a determinant of early adipogenesis. Mol. Cell 62, 443–452 (2016)
Pende, M. et al. Hypoinsulinaemia, glucose intolerance and diminished β-cell size in S6K1-deficient mice. Nature 408, 994–997 (2000)
Taniguchi, C. M., Emanuelli, B. & Kahn, C. R. Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85–96 (2006)
Wiczer, B. M. & Bernlohr, D. A. A novel role for fatty acid transport protein 1 in the regulation of tricarboxylic acid cycle and mitochondrial function in 3T3-L1 adipocytes. J. Lipid Res. 50, 2502–2513 (2009)
Pende, M. et al. S6K1−/−S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24, 3112–3124 (2004)
Ruvinsky, I. et al. Mice deficient in ribosomal protein S6 phosphorylation suffer from muscle weakness that reflects a growth defect and energy deficit. PLoS One 4, e5618 (2009)
Lin, M. H. & Miner, J. H. Fatty acid transport protein 1 can compensate for fatty acid transport protein 4 in the developing mouse epidermis. J. Invest. Dermatol. 135, 462–470 (2015)
Lobo, S., Wiczer, B. M., Smith, A. J., Hall, A. M. & Bernlohr, D. A. Fatty acid metabolism in adipocytes: functional analysis of fatty acid transport proteins 1 and 4. J. Lipid Res. 48, 609–620 (2007)
Coburn, C. T. et al. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J. Biol. Chem. 275, 32523–32529 (2000)
Richards, M. R., Harp, J. D., Ory, D. S. & Schaffer, J. E. Fatty acid transport protein 1 and long-chain acyl coenzyme A synthetase 1 interact in adipocytes. J. Lipid Res. 47, 665–672 (2006)
Ray, P. S. & Fox, P. L. A post-transcriptional pathway represses monocyte VEGF-A expression and angiogenic activity. EMBO J. 26, 3360–3372 (2007)
Kozma, S. C. et al. Active baculovirus recombinant p70s6k and p85s6k produced as a function of the infectious response. J. Biol. Chem. 268, 7134–7138 (1993)
Arif, A., Chatterjee, P., Moodt, R. A. & Fox, P. L. Heterotrimeric GAIT complex drives transcript-selective translation inhibition in murine macrophages. Mol. Cell. Biol. 32, 5046–5055 (2012)
Mazumder, B. et al. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115, 187–198 (2003)
Sampath, P. et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell 119, 195–208 (2004)
Wong, G. W. et al. Molecular, biochemical and functional characterizations of C1q/TNF family members: adipose-tissue-selective expression patterns, regulation by PPAR-gamma agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. Biochem. J. 416, 161–177 (2008)
Lee, R. G. et al. ACAT2 contributes cholesteryl esters to newly secreted VLDL, whereas LCAT adds cholesteryl ester to LDL in mice. J. Lipid Res. 46, 1205–1212 (2005)
Miao, J. et al. Flavin-containing monooxygenase 3 as a potential player in diabetes-associated atherosclerosis. Nat. Commun. 6, 6498 (2015)
Thomas, G. et al. The serine hydrolase ABHD6 is a critical regulator of the metabolic syndrome. Cell Reports 5, 508–520 (2013)
Denzel, M. S. et al. T-cadherin is critical for adiponectin-mediated cardioprotection in mice. J. Clin. Invest. 120, 4342–4352 (2010)
Casnellie, J. E. Assay of protein kinases using peptides with basic residues for phosphocellulose binding. Methods Enzymol. 200, 115–120 (1991)
Kawasome, H. et al. Targeted disruption of p70(s6k) defines its role in protein synthesis and rapamycin sensitivity. Proc. Natl Acad. Sci. USA 95, 5033–5038 (1998)
Burkholder, T., Foltz, C., Karlsson, E., Linton, C. G. & Smith, J. M. Health Evaluation of Experimental Laboratory Mice. Curr. Protoc. Mouse Biol. 2, 145–165 (2012)
Lee, J., Ellis, J. M. & Wolfgang, M. J. Adipose fatty acid oxidation is required for thermogenesis and potentiates oxidative stress-induced inflammation. Cell Reports 10, 266–279 (2015)
Folch, J., Lees, M. & Sloane Stanley, G. H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 (1957)
Kraus, D. et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature 508, 258–262 (2014)
Jia, J., Arif, A., Ray, P. S. & Fox, P. L. WHEP domains direct noncanonical function of glutamyl-prolyl tRNA synthetase in translational control of gene expression. Mol. Cell 29, 679–690 (2008)
Norris, A. W. et al. Muscle-specific PPARγ-deficient mice develop increased adiposity and insulin resistance but respond to thiazolidinediones. J. Clin. Invest. 112, 608–618 (2003)
Mehlem, A., Hagberg, C. E., Muhl, L., Eriksson, U. & Falkevall, A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat. Protocols 8, 1149–1154 (2013)
Liao, J., Sportsman, R., Harris, J. & Stahl, A. Real-time quantification of fatty acid uptake using a novel fluorescence assay. J. Lipid Res. 46, 597–602 (2005)
Carson, S. D. Chromatographic depletion of lipoproteins from plasma and recovery of apolipoproteins. Biochim. Biophys. Acta 750, 317–321 (1983)
Goldstein, J. L., Hoff, H. F., Ho, Y. K., Basu, S. K. & Brown, M. S. Stimulation of cholesteryl ester synthesis in macrophages by extracts of atherosclerotic human aortas and complexes of albumin/cholesteryl esters. Arteriosclerosis 1, 210–226 (1981)
Song, E. K. et al. NAADP mediates insulin-stimulated glucose uptake and insulin sensitization by PPARγ in adipocytes. Cell Reports 2, 1607–1619 (2012)
Chiang, S. H. et al. Insulin-stimulated GLUT4 translocation requires the CAP-dependent activation of TC10. Nature 410, 944–948 (2001)
Acknowledgements
This work was supported by NIH grants P01HL029582, P01HL076491, R01GM086430, R01GM115476, and P50CA150964 (to P.L.F.), by AHA SDG 10SDG3930003 (to A.A), by CIHR fellowship (to D.H), by AHA fellowship (to K.V.), by NIH R01AR048914 and R01GM089771 (to J.C.), by Spanish Ministry BFU2012-38867 and Fundacio La Marato de TV3 #174/U/2016 grants (to S.C.K.), and by NIH R01CA158768, Spanish Ministry SAF2011-24967, and CIG European Commission PCIG10-GA-2011-304160 grants (to G.T.). We thank P. Bhattaram for helpful discussion.
Author information
Authors and Affiliations
Contributions
A.A. and P.L.F. conceived and interpreted most experiments with contributions from J.M.B., J.C., S.C.K., G.T. and X.L. A.A. performed most experiments with contributions from J.J., F.T., J.S., A.C., D.H. and K.V. A.A.P. performed longevity analysis. A.A. and P.L.F. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks A. Stahl and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
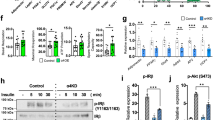
Extended Data Figure 1 Identification of S6K1 as EPRS Ser999 kinase.
a, Screening of AGC kinase group members for phosphorylation of EPRS Ser999 by immunocomplex kinase assay and [γ-32P]ATP-labelling with S886A linker target in U937 cells. Kinase activity using kinase-specific substrate is shown (bottom; mean ± s.e.m., n = 3). b, Specificity of S6K1 for Ser999 phosphorylation, determined by 32P incorporation in wild-type (WT), S999A and S886 linker. c, siRNA targeting S6K1 inhibits IFNγ-stimulated EPRS phosphorylation in U937 cells determined by 32P-labelling (mean ± s.e.m., n = 3). d, Active recombinant kinases used for in vitro phosphorylation of linker bearing S886A (Ser999 P-acceptor) mutation shows site-specific phosphorylation by S6K1. e, Raptor not rictor is required for Ser999 phosphorylation. f, siRNA targeting the S6K1 3′UTR inhibits S6K1 expression and phosphorylation of EPRS Ser999, but not Ser886 (mean ± s.e.m., n = 3). g, Phosphorylation of S6K1 Thr389 is required for phosphorylation of EPRS. Cells were co-transfected with siRNA targeting the 3′-UTR to knock down endogenous S6K1, and with myc-tagged wild-type or mutant S6K1 cDNA; IFNγ-stimulated EPRS phosphorylation determined by 32P-labelling (mean ± s.e.m., n = 3). h, Cells treated as in e but followed by reciprocal co-immunoprecipitation.
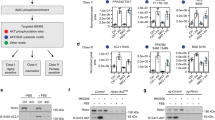
Extended Data Figure 2 Gene targeting and generation of EprsA/A and EprsD/D knock-in mice, and their body weight phenotypes.
a, Knock-in mice bearing Ser999-to-Ala and -Asp mutations were generated by homologous recombination. b, Validation of wild-type (EprsS/S) and EPRS knock-in mice by PCR genotyping (top) and sequence analysis (bottom). c, Genotype analysis of littermates. Total of 1,410 and 644 progeny from interbreeding EprsS/A and EprsS/D mice, respectively, were used at postnatal days P0, P4, and P21. d, Growth curves of EprsA/A (mean ± s.e.m., n = 10 per group; P < 0.0001, two-way ANOVA) and EprsD/D (n = 10 per group) female mice. e, Representative images (left) and weights (right) of wild-type (EprsS/S; S/S) and EprsA/A (A/A) mice on embryonic day E16.5 and post-embryonic development stages. Data shown are mean ± s.e.m.; n = 11 for E16.5 embryos, n = 10 for P0 and P10 mice, and n = 14 for 3, 12, 20, 30, and 50-week mice. f, Representative images (left) and weights (right) of 50-week EprsS/S and EprsD/D (D/D) mice (mean ± s.e.m.; n = 10 per group).
Extended Data Figure 3 Lifespan analysis of EprsS/S, EprsA/A and EprsD/D mice monitored from weaning (>21 days).
a, Youngest and oldest 10% are the mean lifespan of the shortest- and longest-living 10% mice. Numbers of days are represented to nearest full day. Median and mean ± s.e.m.; are shown. b, Cox proportional hazard (CPH) regression analyses of EprsS/S and EprsA/A mice shows genotype as the most significant predictor of increased longevity. Longevity relative to survival days (that is, age at death) was analysed in pooled mice by CPH regression model. The four independent variables genotype, date of birth (DOB), parental ID (PID), and gender were replaced with a set of category variables. In case of genotype and gender, category represents their presence or absence. DOB and PID data were divided into multiple categories as described in the supplementary methods. Independent variables were fitted into the CPH model individually (univariate) or simultaneously (multivariate). Shown are: β (the unstandardized regression coefficient) with standard error (s.e.), the degrees of freedom (df), and the significance for each model fit. Exp(β) for the covariate of interest is the predicted change in hazard ratio for a unit increase in the predictor, and its 95% confidence interval (CI). c, Kaplan–Meier survival curves show no change in lifespan of male, female, or combined EprsD/D mice. Male (MC χ2 = 0.003, P = 0.956; GBW χ2 = 0.001, P = 0.972), female (MC χ2 = 0.158, P = 0. 0.691; GBW χ2 = 0.206, P = 0.650), and gender-combined (MC χ2 = 0.079, P = 0. 0.779; GBW χ2 = 0.076, P = 0.783). d, e, Survival and CPH regression analyses of EprsS/S and EprsD/D mice as described above in a and b.
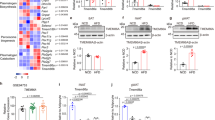
Extended Data Figure 4 Adipose tissue deposition and lipolysis in EPRSA/A and EPRSD/D mice.
a, Length of mice was measured from head to beginning of tail using a digital caliper (Fisherbrand Traceable). Data shown are mean ± s.e.m., n = 15 for 20-week-old male mice. b, Ventral view of wild-type and EprsA/A mice abdominal cavity. c, Weights of adipose and non-adipose tissues from 20-week-old male EprsD/D and control mice (mean ± s.e.m., n = 14 per group, P value from unpaired t-test). d, Scanning electron micrographs of EWAT in 20-week-old male EprsS/S, EprsA/A, and EprsD/D mice. e, Total adipocyte cell number in EWAT of EprsA/A knock-in and wild-type mice. Data represent mean ± s.e.m., n = 5 per group. f, Elevated lipolysis in adipocytes from EprsA/A, but not EprsD/D, mice (mean ± s.e.m.; n = 6 per group). g, Elevated β-oxidation in WAT explants from EprsA/A mice as determined by release of 14CO2 from 14C-oleic acid (mean ± s.e.m.; n = 5 per group). h, Serum levels of insulin, glucose, triglycerides (TG) and free fatty acids (FFA) in 12-h fasted and 1-h post-prandial (fed) 16-week-old male mice (mean ± s.e.m., n = 10 per group, *P < 0.05, unpaired t-test). i, Growth curves of EprsA/A and EprsD/D mice (males, n = 12 per group, mean ± s.e.m., P < 0.001, two-way ANOVA) started at 6 weeks on an unrestricted high-fat diet (Harlan Teklad TD.06414) deriving 18, 60, and 21 kcal% from protein, fat, and carbohydrate, respectively. j, Phosphorylation of EPRS Ser999 in WAT from EprsA/A and EprsD/D mice after high-fat diet feeding for 24 weeks.
Extended Data Figure 5 Adiposity and lifespan of S6K1−/− mice.
a, PCR genotyping of wild-type (S6K1+/+), heterozygous (S6K1+/-) and homozygous (S6K1−/−) mice. b, Immunoblot analysis of S6K1, EPRS, and FATP1 in S6K1−/− mice. c, Weight of S6K1−/− mice at embryonic day E16.5, and at postnatal days P0 and P10. d, Kaplan–Meier survival curves shows increased longevity in male, female or combined S6K1−/− mice. Male (n = 29 per group; MC χ2 = 4.919, P = 0.027; GBW χ2 = 4.660, P = 0.031), female (n = 28 for S6K1+/+ and n = 26 for S6K1−/−; MC χ2 = 7.927, P = 0.005; GBW χ2 = 7.277, P = 0.007), and gender-combined (n = 26 for S6K1+/+ and n = 55 for S6K1−/−; MC χ2 = 11.78, P = 0.0006; GBW χ2 = 11.01, P = 0.0009). e, f, Lifespan and CPH regression analyses of S6K1+/+ and S6K1−/− mice as described above in Extended Data Fig. 3a, b.
Extended Data Figure 6 Glucose metabolism, food intake and faecal lipid excretion in EprsA/A and EprsD/D mice.
a, Glucose tolerance test (GTT) in 112-day-old EprsS/S, EprsA/A, and EprsD/D mice (mean ± s.e.m., n = 10 per group). b, Insulin tolerance test (ITT) on mice as in a (mean ± s.e.m., n = 10 per group). c, d, Same as a and b but using ~600-day-old mice (mean ± s.e.m., n = 9 per group). e–g, Determination of food intake as g per mouse per day (left) or g per body weight (g) per day (right in male (e) and female (f) EprsA/A mice, and in male EprsD/D mice (g). All values represent mean ± s.e.m., n = 14 per group. h, Faecal lipid excretion in EprsS/S, EprsA/A, and EprsD/D mice (mean ± s.e.m., n = 6 per group). i, Serum ketone body (β-hydroxybutyrate) level in 6-h fasted mice.
Extended Data Figure 7 Energy metabolism in EprsA/A and EprsD/D mice.
a, b, Determination of VO2 (left) and VCO2 (right) in EprsS/S and EprsA/A male mice over a 24-h period. c, d, Respiratory exchange ratio (RER) (left) and heat generation (right) in 12-h light and dark cycles were determined (mean ± s.e.m., n = 6 per group). e–h, Same as a–d but comparing EprsS/S and EprsD/D male mice (mean ± s.e.m., n = 6 per group). i, Determination of VO2 in female EprsS/S, EprsA/A, and EprsD/D mice (mean ± s.e.m., n = 3 per group).
Extended Data Figure 8 Absence of GAIT pathway in adipocytes and inflammatory response in EPRSA/A mice.
a, Total protein synthesis determined by [35S]Met/Cys labelling (left), and by incorporation of [14C]Glu and [14C]Pro into TCA-precipitated proteins in adipocytes from EprsS/S, EprsA/A, and EprsD/D mice. b, Effect of siRNA-mediated knockdown of S6K1, raptor, and rictor on phosphorylation of EPRS Ser999 in differentiated mouse 3T3-L1 adipocytes in presence of 100 nM insulin. c, Effect of IFNγ and insulin on phosphorylation of EPRS Ser999 in differentiated 3T3-L1 adipocytes or mouse primary adipocytes determined using phospho-specific EPRS antibody. d, GAIT complex formation in IFNγ-stimulated U937 cells and insulin-stimulated 3T3-L1 adipocytes by immunoprecipitation with anti-EPRS antibody and immunoblot with antibodies against GAIT complex constituents. Cytosolic lysates from IFNγ-treated U937 cells served as positive control. e, Determination of GAIT pathway activity in IFNγ-stimulated U937 cells and insulin-stimulated 3T3-L1 adipocytes by in vitro translation of a control (T7 gene 10) and GAIT element bearing (Luc-ceruloplasmin (Cp) GAIT element) reporter RNAs. f, White blood cells counts in blood from EprsS/S and EprsA/A mice by Advia hematology system (LUC, large unstained cells). g, Determination of cytokine levels in serum from EprsS/S and EprsA/A mice. Mouse cytokine antibody arrays were incubated with serum (100 μg, protein, right). Pixel intensity was determined by densitometry (right; mean ± s.e.m., 2 mice per group). h, Immunoblot analysis of selected cytokines in serum from EprsS/S and EprsA/A mice.
Extended Data Figure 9 Tissue-specificity of insulin-stimulated LCFA uptake and EPRS-FATP1 interaction.
a, [14C]oleate uptake determined in insulin-stimulated hepatocytes, cardiac cells, soleus muscle strips, and BMDM from EprsS/S, EprsA/A, and EprsD/D mice (mean ± s.e.m., n = 6 mice per group). b, Insulin-stimulated EPRS Ser999 phosphorylation (top), [14C]oleate uptake (middle), and [14C]2-deoxy-d-glucose (DG) uptake (bottom) in adipocytes from white adipose tissue of S6K1+/+ and S6K1−/− mice (mean ± s.e.m., n = 5 mice per group). c, Efficiency of EPRS and FATP1 knockdown in 3T3-L1 adipocytes by siRNA targeting each mRNA alone and in combination, as determined by densitometry (NIH image J) of immunoblots shown in Fig. 3g (mean ± s.e.m., n = 4 experiments). d, Insulin-induced EPRS Ser999 phosphorylation, interaction with FATP1, and [14C]oleate uptake in human adipocytes (mean ± s.e.m., n = 3 experiments in duplicate). e, Co-immunoprecipitation experiment to determine FATP1 binding to EPRS in lysates from multiple tissues as indicated. f, EPRS and FATP1 expression in male and female S6K1−/− mice (mean ± s.e.m., n = 3 mice per group). g, Lack of interaction of EPRS and FATP1 in insulin-stimulated adipocytes of S6K1−/− mice.
Extended Data Figure 10 Hepatic lipids and FATP1/EPRS membrane localization in EprsA/A and EprsD/D mice.
a, Optimum cutting temperature (OCT) compound-fixed liver slices from EprsS/S, EprsA/A, and EprsD/D mice were stained with H&E (top) or oil red O (bottom), and the latter quantitated by densitometry (right; mean ± s.e.m., n = 3 mice per group). b, Determination of liver triglycerides in wild-type and mutant mice (mean ± s.e.m., n = 5 mice per group). c, Insulin-inducible membrane localization of EPRS and FATP1 in adipocytes from wild-type and mutant mice. d, Membrane fractionation shows EPRS and FATP1 co-localizing in plasma membrane (mean ± s.e.m., n = 3 experiments).
Supplementary information
Supplementary Figures
This file contains the Source Data gels for the Figures and the Extended Data Figures. (PDF 47832 kb)
Rights and permissions
About this article
Cite this article
Arif, A., Terenzi, F., Potdar, A. et al. EPRS is a critical mTORC1–S6K1 effector that influences adiposity in mice. Nature 542, 357–361 (2017). https://doi.org/10.1038/nature21380
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21380
This article is cited by
-
Inhibition of S6K lowers age-related inflammation and increases lifespan through the endolysosomal system
Nature Aging (2024)
-
ChemRAP uncovers specific mRNA translation regulation via RNA 5′ phospho-methylation
EMBO Reports (2024)
-
A viral pan-end RNA element and host complex define a SARS-CoV-2 regulon
Nature Communications (2023)
-
Targeting the biology of aging with mTOR inhibitors
Nature Aging (2023)
-
Phosphocode-dependent glutamyl-prolyl-tRNA synthetase 1 signaling in immunity, metabolism, and disease
Experimental & Molecular Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.