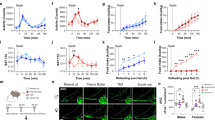
Abstract
Instinctive reactions to danger are critical to the perpetuation of species and are observed throughout the animal kingdom. The scent of predators induces an instinctive fear response in mice that includes behavioural changes, as well as a surge in blood stress hormones that mobilizes multiple body systems to escape impending danger1,2. How the olfactory system routes predator signals detected in the nose to achieve these effects is unknown. Here we identify a specific area of the olfactory cortex in mice that induces stress hormone responses to volatile predator odours. Using monosynaptic and polysynaptic viral tracers, we found that multiple olfactory cortical areas transmit signals to hypothalamic corticotropin-releasing hormone (CRH) neurons, which control stress hormone levels. However, only one minor cortical area, the amygdalo-piriform transition area (AmPir), contained neurons upstream of CRH neurons that were activated by volatile predator odours. Chemogenetic stimulation of AmPir activated CRH neurons and induced an increase in blood stress hormones, mimicking an instinctive fear response. Moreover, chemogenetic silencing of AmPir markedly reduced the stress hormone response to predator odours without affecting a fear behaviour. These findings suggest that AmPir, a small area comprising <5% of the olfactory cortex, plays a key part in the hormonal component of the instinctive fear response to volatile predator scents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Apfelbach, R., Blanchard, C. D., Blanchard, R. J., Hayes, R. A. & McGregor, I. S. The effects of predator odors in mammalian prey species: a review of field and laboratory studies. Neurosci. Biobehav. Rev. 29, 1123–1144 (2005)
Takahashi, L. K. Olfactory systems and neural circuits that modulate predator odor fear. Front. Behav. Neurosci. 8, 72 (2014)
Ulrich-Lai, Y. M. & Herman, J. P. Neural regulation of endocrine and autonomic stress responses. Nature Rev. Neurosci. 10, 397–409 (2009)
Kobayakawa, K. et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature 450, 503–508 (2007)
Papes, F., Logan, D. W. & Stowers, L. The vomeronasal organ mediates interspecies defensive behaviors through detection of protein pheromone homologs. Cell 141, 692–703 (2010)
Buck, L. B. & Bargmann, C. in Principles of Neuroscience (eds Kandel, E., Schwartz, J., Jessell, T., Siegelbaum, S. & Hudspeth, A. J. ) 712–742 (McGraw-Hill, 2012)
Neville, K. R. & Haberly, L. B. in The Synaptic Organization of the Brain (ed. Shepherd, G. M. ) 415–454 (Oxford University Press, 2004)
Buck, L. & Axel, R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187 (1991)
Ressler, K. J., Sullivan, S. L. & Buck, L. B. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73, 597–609 (1993)
Vassar, R., Ngai, J. & Axel, R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74, 309–318 (1993)
Hanchate, N. K. et al. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science 350, 1251–1255 (2015)
Ressler, K. J., Sullivan, S. L. & Buck, L. B. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell 79, 1245–1255 (1994)
Vassar, R. et al. Topographic organization of sensory projections to the olfactory bulb. Cell 79, 981–991 (1994)
Mombaerts, P. et al. Visualizing an olfactory sensory map. Cell 87, 675–686 (1996)
Sosulski, D. L., Bloom, M. L., Cutforth, T., Axel, R. & Datta, S. R. Distinct representations of olfactory information in different cortical centres. Nature 472, 213–216 (2011)
Miyamichi, K. et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature 472, 191–196 (2011)
Ghosh, S. et al. Sensory maps in the olfactory cortex defined by long-range viral tracing of single neurons. Nature 472, 217–220 (2011)
Root, C. M., Denny, C. A., Hen, R. & Axel, R. The participation of cortical amygdala in innate, odour-driven behaviour. Nature 515, 269–273 (2014)
Card, J. P. & Enquist, L. W. Transneuronal circuit analysis with pseudorabies viruses. Curr. Protoc. Neurosci. Chapter 1, Unit 1.5 (2001)
DeFalco, J. et al. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science 291, 2608–2613 (2001)
Krashes, M. J. et al. An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507, 238–242 (2014)
Sawchenko, P. E. & Swanson, L. W. The organization of forebrain afferents to the paraventricular and supraoptic nuclei of the rat. J. Comp. Neurol. 218, 121–144 (1983)
Guzowski, J. F., McNaughton, B. L., Barnes, C. A. & Worley, P. F. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nature Neurosci. 2, 1120–1124 (1999)
Vernet-Maury, E. in Olfaction and Taste (ed. Van der Starre, H. ) 407 (IRL, 1980)
Krashes, M. J. et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 121, 1424–1428 (2011)
Gorski, J. A. et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314 (2002)
Vong, L. et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71, 142–154 (2011)
Armbruster, B. N., Li, X., Pausch, M. H., Herlitze, S. & Roth, B. L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl Acad. Sci. USA 104, 5163–5168 (2007)
Matsukawa, M., Imada, M., Murakami, T., Aizawa, S. & Sato, T. Rose odor can innately counteract predator odor. Brain Res. 1381, 117–123 (2011)
Franklin, K. & Paxinos, G. The Mouse Brain in Stereotaxic Coordinates 3rd edn (Academic, 2008)
Schnütgen, F. et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nature Biotechnol. 21, 562–565 (2003)
Banfield, B. W. & Bird, G. A. Construction and analysis of alphaherpesviruses expressing green fluorescent protein. Methods Mol. Biol. 515, 227–238 (2009)
Tiscornia, G., Singer, O. & Verma, I. M. Production and purification of lentiviral vectors. Nature Protocols 1, 241–245 (2006)
Boehm, U., Zou, Z. & Buck, L. B. Feedback loops link odor and pheromone signaling with reproduction. Cell 123, 683–695 (2005)
Liberles, S. D. & Buck, L. B. A second class of chemosensory receptors in the olfactory epithelium. Nature 442, 645–650 (2006)
Acknowledgements
We thank L. Enquist for providing the parental PRV, PRV TK-BaBlu, and for helpful advice, and members of the Buck laboratory for discussions and comments. This work was supported by the Howard Hughes Medical Institute (HHMI) and by grants from the National Institutes of Health (National Institute on Deafness and Other Communication Disorders) (L.B.B.) and the Japan Society for the Promotion of Science (K.K.). L.B.B. is an Investigator of the HHMI.
Author information
Authors and Affiliations
Contributions
K.K. and L.B.B. conceived the project, Z.L. and K.K. designed and prepared the viruses, X.Y. performed experiments, D.P.O. and B.B.L. generated the CRH-Cre mice, K.K. developed methods and performed most of the experiments and data analysis, and K.K. and L.B.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Virus infection and spread in CRH-Cre and wild-type mice.
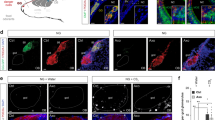
a, Photographs show a PVN section from CRH-Cre mice crossed with Rosa-floxstop-GFP mice that was co-stained with anti-GFP antibodies (green) (Cre) and a Crh riboprobe (red) (Crh). Co-labelled cells (yellow) can be seen in the merged image, verifying the expression of Cre in CRH neurons. Of GFP+ (Cre+) neurons in PVN sections, 93.9 ± 0.5% expressed Crh. n = 4. Scale bar, 100 μm. b, Photographs show a section through the PVN of a CRH-Cre mouse previously injected with PRVB316 and LVF2TK. The section was co-stained with antibodies for Cre-dependent reporters of PRVB316 (GFP (green)) and LVF2TK (HA (red)). Some co-labelled neurons are seen (arrows), verifying co-infection of some neurons by the two viruses. Scale bar, 200 μm. c, The number of PRVB316+ cells inside or outside the PVN, 4 days after PVN injection of PRVB316 alone (n = 2) or PRVB316+LVF2TK (n = 24) in CRH-Cre mice or PRVB316+LVF2TK in wild-type (WT) mice (n = 3). Error bars indicate s.e.m. In CRH-Cre mice, PRVB316+ cells were seen both inside and outside the PVN after co-injection of PRVB316 and LVF2TK, but only inside the PVN when only PRVB316 was injected. In wild-type mice co-injected with the two viruses, PRV+ cells were not detected either inside or outside the PVN. d, The number of PRV+ cells inside or outside the PVN 3 or 5 days after PVN injection of PRVB177 in CRH-Cre or wild-type mice. n = 8 (CRH-Cre mice, day 3), n = 6 (CRH-Cre mice, day 5), n = 2 (wild-type mice, day 3), n = 2 (wild-type mice, day 5). Error bars indicate s.e.m. PRVB177+ neurons were seen both inside and outside the PVN of CRH-Cre mice on both days, but were not detected in wild-type mice at either location.
Extended Data Figure 2 Brain areas with neurons upstream of CRH neurons.
The percentage of all non-PVN PRV+ neurons found in individual brain areas 4 days after PVN injection of the monosynaptic PRV (PRVB316; Mono) (n = 24) or on day 3, 4 or 5 after PVN injection of the polysynaptic PRV (PRVB177; Poly) (n = 8, 6 and 6, for day 3, 4 and 5, respectively). Error bars indicate s.e.m. See Methods for full names of abbreviated brain areas.
Extended Data Figure 3 Brain areas containing neurons upstream of CRH neurons.
Coloured boxes indicate the approximate number of infected neurons seen in individual brain areas (white indicates none) on day 4 after PVN injection of CRH-Cre mice with the monosynaptic PRV (PRVB316; Mono) (n = 24) or on day 3, 4 or 5 after injection with the polysynaptic PRV (PRVB177; Poly) (n = 8, 6, and 6, for day 3, 4 and 5, respectively). See Methods for full names of abbreviated brain areas.
Extended Data Figure 4 Biased distribution of neurons upstream of CRH neurons in the pPir, AmPir and MEA.
a, b, Diagrams show the numbers of PRVB177-infected (HA+) neurons on day 4 after PVN injection in sequential 20-μm coronal sections along the anterior (A)–posterior (P) axis in different areas of the OC (a) and VA (b). Data are shown in separate panels for four animals for each area. Infected neurons appear to be randomly distributed along the anterior–posterior axis in some areas, but not others. c–e, Photographs of sections immunostained for PRVB177 (HA, green) on day 4 after PVN injection of PRVB177. Sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). More PRVB177+ neurons are seen in the posterior than anterior pPir (c) and AmPir (d). In the MEA (e), more are seen in the posteroventral (pv) quadrant than other quadrants (posterodorsal (pd), anterodorsal (ad), and anteroventral (av)). Scale bars, 200 μm. f–h, The percentages of PRVB177-infected neurons in the anterior versus posterior halves of the pPir (f) and AmPir (g) and in different quadrants of the MEA (h). n = 4. Error bars indicate s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. Paired t-test (f, g) and paired one-way analysis of variance (ANOVA) with post-hoc Tukey’s test (h). Infected neurons were concentrated in the posterior halves of the pPir and AmPir and were more abundant in the posteroventral quadrant of the MEA than in other quadrants.
Extended Data Figure 5 Expression of glutamatergic and GABAergic markers in PRV-infected neurons.
a–c, Photographs of sections through the pPir (a), AmPir (b) or MEAp (c) 4 days after PVN injection of PRVB177. Sections were co-stained with antibodies against HA (PRVB177) (green) and riboprobes (red) for Vglut1 and Vglut2 (Vglut1/2), to identify glutamatergic neurons (Glu), or Gad1 and Gad2 (Gad1/2), to identify GABAergic neurons (GABA). Sections were counterstained with DAPI. Arrowheads indicate co-labelled neurons. Numerous PRV-infected neurons were co-labelled for Vglut1/2 in all three areas, but only the MEAp contained many co-labelled for Gad1/2. Scale bars, 100 μm. d, The percentage of PRVB177+ neurons co-labelled for glutamatergic (Vglut1/2+) or GABAergic (Gad1/2+) markers in different areas of the OC and VA. PRV+ neurons unlabelled by either probe could express other neurotransmitters or neuromodulators or reflect PRV effects on RNA transcription or degradation. n = 4 mice per condition. Error bars indicate s.e.m. Most of the co-labelled PRV+ cells in the olfactory areas were co-labelled for the glutamatergic makers, but the MEA also contained some co-labelled for the GABAergic markers.
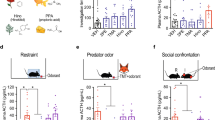
Extended Data Figure 6 TMT and bobcat urine activate CRH neurons and induce stress hormone increases.
a, Photographs of PVN sections from CRH-Cre mice crossed with Rosa-floxstop-GFP mice. Mice were exposed to water (H2O), rabbit urine, bobcat urine or TMT and sections were then co-stained with antibodies against GFP (green), to identify CRH neurons, and c-Fos (red), to detect activated neurons. Higher magnifications of boxed areas are shown on the right. Many CRH neurons (GFP+) expressed c-Fos after exposure to bobcat urine or TMT, but not rabbit urine. Scale bars, 100 μm (left) and 20 μm (right). b, The percentage of PVN CRH (GFP+) neurons immunostained for c-Fos after odour exposure of CRH-Cre mice crossed with Rosa-floxstop-GFP mice. Exposure to bobcat urine or TMT, but not rabbit urine, increased the percentage of CRH neurons expressing c-Fos. n = 6 mice per condition. Error bars indicate s.e.m. ***P < 0.001, one-way ANOVA with post-hoc Dunnett’s test. c, d, Plasma ACTH (c) or corticosterone (d) concentrations were measured in wild-type mice after odour exposure. Exposure of animals to TMT or bobcat urine increased blood levels of ACTH and corticosterone, whereas exposure to rabbit urine did not. n = 6 mice per condition. Error bars indicate s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA with post-hoc Dunnett’s test (c) or Kruskal–Wallis test with post-hoc Dunn’s test (d).
Extended Data Figure 7 Locations of infected neurons after injection of AAV-DIO-hM3Dq-mCherry.
a–c, AAV-DIO-hM3Dq-mCherry was injected into the AmPir (a), pPir (b) or MEA (c) and brain sections immunostained for mCherry to detect infected neurons. Shown are schematic sections from a mouse brain atlas30 that correspond to immunostained sections. The target area (AmPir, pPir or MEA) is indicated in purple for each section. Regions that contained the nuclei of mCherry+ neurons are indicated in red. Schematic sections are shown for six mice per area that were injected with CNO to test the effects of activating the area.
Extended Data Figure 8 Locations of infected neurons after injection of AAV-DIO-hM4Di-mCherry.
a–c, AAV-DIO-hM4Di-mCherry was injected into the AmPir (a, b) or PLCo/PMCo (c) and brain sections immunostained for mCherry to detect infected neurons. Shown are schematic sections from a mouse brain atlas30 that correspond to immunostained sections. The location of the AmPir is indicated in purple (a–c), and the location of the PLCo/PMCo is indicated in green (c). Regions that contained the nuclei of mCherry+ neurons are indicated in red in each section. Schematics are shown for six mice per area (per condition) injected with CNO and then exposed to bobcat urine (a, c) or TMT (b) to test the effects of silencing the AmPir or PLCo/PMCo.
Extended Data Figure 9 Effect of neuronal silencing on the activation of neurons by predator odours.
a–c, AAV-DIO-hM4Di-mCherry was injected into the AmPir (a, b) or PLCo/PMCo (c). Animals were later injected with CNO or saline and exposed to bobcat urine (a, c) or TMT (b). Brain sections were hybridized with an Arc riboprobe and the number of cells labelled for nArc was determined for the AmPir, PLCo and PMCo. Histograms show the number of cells with nArc in each area after injection with CNO or saline and exposure to predator odour. On the basis of the numbers of nArc+ neurons, silencing of AmPir neurons caused a significant decrease in AmPir and PLCo neurons activated by bobcat urine (a) and by AmPir (but not PLCo) neurons activated by TMT (b). Silencing of the PLCo/PMCo (c) caused a significant decrease in neurons activated by bobcat urine in the PLCo, but not the AmPir or PMCo. n = 6–7 per condition. Error bars indicate s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired t-test.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1. (PDF 133 kb)
Rights and permissions
About this article
Cite this article
Kondoh, K., Lu, Z., Ye, X. et al. A specific area of olfactory cortex involved in stress hormone responses to predator odours. Nature 532, 103–106 (2016). https://doi.org/10.1038/nature17156
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17156
This article is cited by
-
A Novel Retrograde AAV Variant for Functional Manipulation of Cortical Projection Neurons in Mice and Monkeys
Neuroscience Bulletin (2024)
-
Olfactory modulation of stress-response neural circuits
Experimental & Molecular Medicine (2023)
-
Brain-wide perception of the emotional valence of light is regulated by distinct hypothalamic neurons
Molecular Psychiatry (2022)
-
Targeting thalamic circuits rescues motor and mood deficits in PD mice
Nature (2022)
-
Odor blocking of stress hormone responses
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.