Abstract
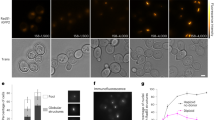
Escherichia coli RecA is the defining member of a ubiquitous class of DNA strand-exchange proteins that are essential for homologous recombination, a pathway that maintains genomic integrity by repairing broken DNA1. To function, filaments of RecA must nucleate and grow on single-stranded DNA (ssDNA) in direct competition with ssDNA-binding protein (SSB), which rapidly binds and continuously sequesters ssDNA, kinetically blocking RecA assembly2,3. This dynamic self-assembly on a DNA lattice, in competition with another protein, is unique for the RecA family compared to other filament-forming proteins such as actin and tubulin. The complexity of this process has hindered our understanding of RecA filament assembly because ensemble measurements cannot reliably distinguish between the nucleation and growth phases, despite extensive and diverse attempts2,3,4,5. Previous single-molecule assays have measured the nucleation and growth of RecA—and its eukaryotic homologue RAD51—on naked double-stranded DNA and ssDNA6,7,8,9,10,11,12; however, the template for RecA self-assembly in vivo is SSB-coated ssDNA3. Using single-molecule microscopy, here we directly visualize RecA filament assembly on single molecules of SSB-coated ssDNA, simultaneously measuring nucleation and growth. We establish that a dimer of RecA is required for nucleation, followed by growth of the filament through monomer addition, consistent with the finding that nucleation, but not growth, is modulated by nucleotide and magnesium ion cofactors. Filament growth is bidirectional, albeit faster in the 5′→3′ direction. Both nucleation and growth are repressed at physiological conditions, highlighting the essential role of recombination mediators in potentiating assembly in vivo. We define a two-step kinetic mechanism in which RecA nucleates on transiently exposed ssDNA during SSB sliding and/or partial dissociation (DNA unwrapping) and then the RecA filament grows. We further demonstrate that the recombination mediator protein pair, RecOR (RecO and RecR), accelerates both RecA nucleation and filament growth, and that the introduction of RecF further stimulates RecA nucleation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kowalczykowski, S. C. & Eggleston, A. K. Homologous pairing and DNA strand-exchange proteins. Annu. Rev. Biochem. 63, 991–1043 (1994)
Kowalczykowski, S. C. Biochemistry of genetic recombination: energetics and mechanism of DNA strand exchange. Annu. Rev. Biophys. Biophys. Chem. 20, 539–575 (1991)
Kowalczykowski, S. C., Clow, J., Somani, R. & Varghese, A. Effects of the Escherichia coli SSB protein on the binding of Escherichia coli RecA protein to single-stranded DNA: demonstration of competitive binding and the lack of a specific protein-protein interaction. J. Mol. Biol. 193, 81–95 (1987)
Thresher, R. J., Christiansen, G. & Griffith, J. D. Assembly of presynaptic filaments. Factors affecting the assembly of RecA protein onto single-stranded DNA. J. Mol. Biol. 201, 101–113 (1988)
Cazenave, C., Toulme, J. J. & Helene, C. Binding of RecA protein to single-stranded nucleic acids: spectroscopic studies using fluorescent polynucleotides. EMBO J. 2, 2247–2251 (1983)
Shivashankar, G. V., Feingold, M., Krichevsky, O. & Libchaber, A. RecA polymerization on double-stranded DNA by using single-molecule manipulation: the role of ATP hydrolysis. Proc. Natl Acad. Sci. USA 96, 7916–7921 (1999)
Galletto, R., Amitani, I., Baskin, R. J. & Kowalczykowski, S. C. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 443, 875–878 (2006)
Joo, C. et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell 126, 515–527 (2006)
Handa, N., Amitani, I., Gumlaw, N., Sandler, S. J. & Kowalczykowski, S. C. Single molecule analysis of a red fluorescent RecA protein reveals a defect in nucleoprotein filament nucleation that relates to its reduced biological functions. J. Biol. Chem. 284, 18664–18673 (2009)
Modesti, M. et al. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure 15, 599–609 (2007)
van der Heijden, T. et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 35, 5646–5657 (2007)
Hilario, J., Amitani, I., Baskin, R. J. & Kowalczykowski, S. C. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl Acad. Sci. USA 106, 361–368 (2009)
Dillingham, M. S. et al. Fluorescent single-stranded DNA binding protein as a probe for sensitive, real-time assays of helicase activity. Biophys. J. 95, 3330–3339 (2008)
Bell, J. C. Dynamic Assembly of RecA Filaments on Single Molecules of SSB-coated ssDNA. PhD thesis, Univ. of California, Davis. (2011)
Griffith, J. D., Harris, L. D. & Register, J., III Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harb. Symp. Quant. Biol. 49, 553–559 (1984)
Kowalczykowski, S. & Steinhardt, J. Kinetics of hemoglobin S gelation followed by continuously sensitive low-shear viscosity. J. Mol. Biol. 115, 201–213 (1977)
Chen, Z., Yang, H. & Pavletich, N. P. Mechanism of homologous recombination from the RecA–ssDNA/dsDNA structures. Nature 453, 489–494 (2008)
Berg, O. G. & von Hippel, P. H. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 14, 131–158 (1985)
Register, J. C., III & Griffith, J. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J. Biol. Chem. 260, 12308–12312 (1985)
Menetski, J. P. & Kowalczykowski, S. C. Enhancement of Escherichia coli RecA protein enzymatic function by dATP. Biochemistry 28, 5871–5881 (1989)
Menetski, J. P. & Kowalczykowski, S. C. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 181, 281–295 (1985)
McEntee, K., Weinstock, G. M. & Lehman, I. R. Binding of the recA protein of Escherichia coli to single- and double-stranded DNA. J. Biol. Chem. 256, 8835–8844 (1981)
Umezu, K., Chi, N. W. & Kolodner, R. D. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl Acad. Sci. USA 90, 3875–3879 (1993)
Morimatsu, K. & Kowalczykowski, S. C. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11, 1337–1347 (2003)
Handa, N., Morimatsu, K., Lovett, S. T. & Kowalczykowski, S. C. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev. 23, 1234–1245 (2009)
Forget, A. L. & Kowalczykowski, S. C. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature 482, 423–427 (2012)
Lohman, T. M. & Kowalczykowski, S. C. Kinetics and mechanism of the association of the bacteriophage T4 gene 32 (helix destabilizing) protein with single-stranded nucleic acids: evidence for protein translocation. J. Mol. Biol. 152, 67–109 (1981)
Roy, R., Kozlov, A. G., Lohman, T. M. & Ha, T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 461, 1092–1097 (2009)
Kuznetsov, S. V., Kozlov, A. G., Lohman, T. M. & Ansari, A. Microsecond dynamics of protein-DNA interactions: direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. J. Mol. Biol. 359, 55–65 (2006)
Umezu, K. & Kolodner, R. D. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem. 269, 30005–30013 (1994)
Kowalczykowski, S. C. & Krupp, R. A. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein: evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J. Mol. Biol. 193, 97–113 (1987)
Napolitano, M. A. et al. Brg1 chromatin remodeling factor is involved in cell growth arrest, apoptosis and senescence of rat mesenchymal stem cells. J. Cell Sci. 120, 2904–2911 (2007)
Hamon, L. et al. High-resolution AFM imaging of single-stranded DNA-binding (SSB) protein–DNA complexes. Nucleic Acids Res. 35, e58 (2007)
Plank, J. L. & Hsieh, T. S. A novel, topologically constrained DNA molecule containing a double Holliday junction: design, synthesis, and initial biochemical characterization. J. Biol. Chem. 281, 17510–17516 (2006)
Thorpe, H. M. & Smith, M. C. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl Acad. Sci. USA 95, 5505–5510 (1998)
Acknowledgements
We are grateful to members of the laboratory for their comments on this work. J.C.B. was funded by a National Institutes of Health (NIH) Predoctoral Training Program in Molecular & Cellular Biology (T32 GM007377); C.C.D. and J.L.P. were funded by the NIH T32 Training Program in Oncogenic Signals and Chromosome Biology (CA10052159); J.L.P. was also funded by a NIH Postdoctoral Fellowship (CA136103); and S.C.K. was supported by the NIH (GM62653 and GM64745).
Author information
Authors and Affiliations
Contributions
J.C.B., J.L.P., C.C.D. and S.C.K. conceived the general ideas, designed the experiments and interpreted the data. J.C.B., J.L.P. and C.C.D. performed experiments. J.C.B. and S.C.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1, Supplementary Figures 1-6 and full legends for Supplementary Videos 1 and 2. (PDF 2723 kb)
Direct imaging of nucleation and growth of RecA on SSB-coated ssDNA
Video illustrating the procedure and showing direct imaging of nucleation and growth of RecA on SSB-coated ssDNA. See Supplementary Information file for full legend. (MOV 3132 kb)
Optical trapping and manipulation of single molecules of gapped DNA for direct imaging of RecAf filament assembly
Video illustrating the procedure and showing optical trapping and manipulation of single molecules of gapped DNA for direct imaging of RecAf filament assembly. See Supplementary Information file for full legend. (MOV 9278 kb)
Rights and permissions
About this article
Cite this article
Bell, J., Plank, J., Dombrowski, C. et al. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature 491, 274–278 (2012). https://doi.org/10.1038/nature11598
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11598
This article is cited by
-
An automated single-molecule FRET platform for high-content, multiwell plate screening of biomolecular conformations and dynamics
Nature Communications (2023)
-
Compartmentalization of the replication fork by single-stranded DNA-binding protein regulates translesion synthesis
Nature Structural & Molecular Biology (2022)
-
Optical tweezers in single-molecule biophysics
Nature Reviews Methods Primers (2021)
-
Antibiotic-induced DNA damage results in a controlled loss of pH homeostasis and genome instability
Scientific Reports (2020)
-
A change of view: homologous recombination at single-molecule resolution
Nature Reviews Genetics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.