The cells that circulate in the bloodstream perform various functions and, in adults, are derived from progenitor cells in the bone marrow. Mutations in the DNA sequences of progenitor cells can lead to changes in blood-cell development, sometimes resulting in cancer. Owing to technical constraints, elucidating the effects of progenitor mutations on blood-cell development has been challenging. Writing in Nature, Nam et al.1 report a method for detecting mutations and measuring gene expression in individual blood progenitor cells, and use it to analyse a mixture of progenitors with or without mutations in a cancer-linked gene. They show that progenitors that have the same mutation can give rise to cells with different gene-expression profiles.
Read the paper: Somatic mutations and cell identity linked by Genotyping of Transcriptomes
Haematopoiesis — the process through which mature blood cells are formed from progenitors — is tightly regulated. The ‘decision’ that progenitor cells make as to which cell type to become is generally determined by the signals that they receive from their immediate surroundings. However, mutations that sometimes arise in these progenitor cells can result in the signals being blocked, over-amplified or simply ignored, resulting in the enrichment or depletion of specific cell types and, in some cases, production of cancerous clones. Understanding how mutations in progenitor cells lead to changes in the production of different cell types is a key question.
Investigating how mutations in a progenitor cell affect its gene expression, and thus its identity and function, has been highly challenging, largely because mutant cells can be rare and often do not express molecular markers that can be used to separate them physically from non-mutant cells. Strategies to simultaneously detect genetic differences and measure gene expression in single cells have been used to assign cells from a mixture of immune blood cells to their human donor of origin2, and to study changes in populations of host and donor cells in individuals with a type of blood cancer who received stem-cell transplants3. However, combined approaches have not been extensively used to examine the effects of mutations in cancer-associated genes on blood-cell development.
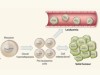
Nam et al. designed a method called ‘genotyping of transcriptomes’ (GoT) by combining an existing platform for profiling gene expression3 with a technique for amplifying a specific genetic sequence to detect mutations in it (Fig. 1). They used this method to analyse thousands of progenitor cells sampled from the bone marrow of five individuals with a form of blood cancer that is caused by mutations in the CALR gene, and that is characterized by overproduction of platelet cells. GoT enabled the authors to ascertain which of the sampled cells carried a CALR mutation and which did not.

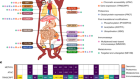
Figure 1 | An analysis of mutation status and gene expression in single cells. Nam et al.1 sampled progenitor cells that give rise to blood cells from individuals who have a type of blood cancer that is caused by progenitor cells with mutations in the CALR gene. To distinguish mutant from non-mutant cells, the authors amplified and sequenced the CALR gene of individual cells. The authors also measured the levels of gene expression in each cell. They identified different cell types on the basis of a statistical analysis of the cells’ gene-expression profiles (dotted circles represent statistical, rather than physical, cell groupings), and examined which of the cells in these different types had CALR mutations. Certain cell types were enriched in CALR-mutant cells, and CALR mutations had different effects (for example, on proliferation) in cells of different types.
The authors used a statistical analysis to ‘group’ the sampled progenitor cells into different types on the basis of their gene-expression profiles (Fig. 1). All of the identified types contained both cells with and without the CALR mutation. However, CALR-mutant cells were more likely to follow certain differentiation pathways and therefore to become certain types of blood cell. Furthermore, Nam and colleagues found that the effects of the mutation, when present in the progenitor cells, were noticeable only at later stages of cellular differentiation; the progeny of CALR-mutant cells were more abundant than the progeny of their non-mutant counterparts and had a distinct gene-expression profile. Such observations would not have been possible using standard techniques, which demonstrates the value of this method.
Although GoT has its limitations, they can probably be addressed by adapting it to new single-cell workflows. First, GoT currently requires that the identity of the mutated gene, or a small set of potentially mutated genes, is known in advance. As an example, the authors used a multiplexed version of their analysis that can simultaneously target multiple prespecified parts of the genetic sequence to probe three genes. If no specific mutations, genes or regions of the genome have been prespecified for analysis (for example, on the basis of an association with disease progression), multiplexed analyses can, in theory, be used to cover larger panels of genes; however, this might not be cost-effective.
Second, GoT is less effective at detecting mutations that occur near the middle of a gene than those that occur near the ends. One solution to this problem would be to use a lower-throughput platform that allows the analysis of full-length RNA transcripts in single cells4,5; in theory, this approach could detect mutations anywhere in the RNA-encoding parts of genes. Nam et al. present an alternative approach by showing that a technique called nanopore sequencing, in which full-length transcripts are sequenced by passing them through a tiny pore, is compatible with their high-throughput platform.
Third, GoT cannot detect mutations in genetic sequences that are not transcribed but that may affect gene expression. Investigation of such sequences might be possible by combining GoT with a technique that measures how accessible certain DNA sequences in a cell are to enzymes6.
A recent paper7 used a different high-throughput approach to implement a similar targeted-amplification strategy to study a blood cancer that is thought to be partly caused by disruption of haematopoiesis by progenitor-cell mutations. The authors of that paper also identified a set of genes that were co-expressed only in malignant progenitors (that is, progenitor cells with a cancer-associated mutation), and described a machine-learning approach that used gene-expression data to distinguish malignant cells from non-malignant ones, even without using prespecified gene-sequence information. It would be interesting to see whether the same machine-learning approach could use Nam and colleagues’ gene-expression data to distinguish the malignant cells from non-malignant cells. Obtaining gene-sequence information from single cells remains more challenging than assessing gene expression; therefore, a method for predicting malignancy solely on the basis of single-cell gene expression would have vast clinical implications.
In theory, GoT and similar approaches could be used to study any cancer. They have the potential to precisely determine the effects of mutations in known genes on downstream cell-development states and to establish whether certain mutations are sufficient to induce cancer. These insights, in turn, could shed light on the mechanisms that underlie the evolution of clonal lineages of cells in cancer.

 Read the paper: Somatic mutations and cell identity linked by Genotyping of Transcriptomes
Read the paper: Somatic mutations and cell identity linked by Genotyping of Transcriptomes
 Genetic clues can be used to predict whether early-stage cancer will form an invasive tumour
Genetic clues can be used to predict whether early-stage cancer will form an invasive tumour
 Bad blood promotes tumour progression
Bad blood promotes tumour progression