Abstract
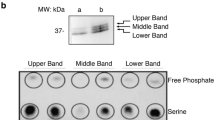
Protein-tyrosine kinases, either in the form of growth-factor receptors1–3 or as the polypeptide products of oncogenes4,5, appear to be important in the regulation of cell growth and transformation. A major question, however, is how their substrates mediate changes in gene expression (reviewed in refs 6–8). Because binding of proteins to specific DNA sequences represents the most direct mechanism for regulating transcription, we have investigated the possibility that some DNA-binding proteins may be substrates of protein-tyrosine kinases. Here, we present evidence for nuclear phosphotyrosyl-proteins in murine fibroblasts transformed by the v-abl protein-tyrosine kinase. Furthermore, we have found that these proteins are not significantly phosphorylated in normal NIH 3T3 cells. Finally, using affinity competition chromatography with bacterial and mouse DNA, we have demonstrated that some of these proteins preferentially bind to mouse DNA. The identification of phosphotyrosyl-proteins with selective DNA-binding properties suggests a possible mechanism through which protein-tyrosine kinases may effect changes in gene transcription.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Ushiro, H. & Cohen, S. J. biol. Chem. 255, 8363–8365 (1980).
Ek, B., Westermark, B., Wasteson, A. & Heldin, C. H. Nature 295, 419–420 (1982).
Scherr, C. J. et al. Cell 41, 665–676 (1985).
Hunter, T. & Sefton, B. M. Proc. natn. Acad. Sci. U.S.A. 77, 1311–1315 (1980).
Witte, O. N., Dasgupta, A. & Baltimore, D. Nature 283, 826–831 (1980).
Zullo, J. N., Cochran, B. H., Huang, A. S. & Stiles, C. D. Cell 43, 793–800 (1985).
Hunter, T. & Cooper, J. A. A. Rev. Biochem. 54, 897–930 (1985).
Foulkes, J. G. & Rich Rosner, M. in Molecular Aspects of Cellular Regulation 4 (eds Cohen, P. & Houslay, M.) 217–252 (Elsevier, Amsterdam, 1985).
Scher, C. D. & Siegler, R. Nature 253, 729–731 (1975).
Frackelton, A. R., Ross, A. H. & Eisen, N. N. Molec. Cell Biol. 3, 3173–3178 (1984).
Sanzo, M., Stevens, B., Tsai, M. & O'Malley, B. W. Biochemistry 23, 6491–6498 (1984).
Tse-Dinh, Y. C., Wong, T. W. & Goldberg, A. R. Nature 312, 785–786 (1984).
Blat, C., Harel, L., Villaudy, J. & Golde, A. Expl Cell Res. 134, 121–128 (1981).
Blat, C., Harel, L., Villaudy, J. & Golde, A. Expl Cell Res. 145, 305–314 (1983).
Ghosh-Dastidar, P., Coty, W. A., Griest, R. E., Woo, D. D. L. & Fox, C. F. Proc. natn. Acad. Sci. U.S.A. 81, 1645–1658 (1984).
Migliaccio, A., Rotondi, A. & Auricchio, F. Proc. natn. Acad. Sci. U.S.A. 81, 5921–5925 (1984).
Colledge, W. H., Edge, M. & Foulkes, J. G. Biosc. Rep. 6, 301–307 (1986).
Migliaccio, A., Rotondi, A. & Auricchio, F. EMBO J. 5, 2867–2872 (1986).
Miller, K., Beardmore, J., Kanety, H., Schlessinger, J. & Hopkins, C. R. J. Cell Biol. 102, 500–509 (1986).
Resh, M. D. & Erikson, R. L. J. Cell Biol. 100, 409–417 (1985).
Cabaud, P. G. & Wroblewski, F. Am. J. clin. Path. 30, 234 (1958).
Newby, A. C., Luzio, J. P. & Hales, C. N. Biochem. J. 146, 625–633 (1975).
Beufay, H. et al. J. Cell Biol. 61, 213–231 (1974).
Bretz, R. & Staubli, W. Eur. J. Bioch. 77, 181–192 (1977).
Cooperstein, S. J. & Lazarow, A. J. biol. Chem. 189, 665–670 (1951).
Labarca, C. & Paigen, K. Analyt. Biochem. 102, 344–352 (1980).
Heath, J. K., Mahadevan, L. & Foulkes, J. G. EMBO J. 5, 1809–1814 (1986).
O'Farrell, P. H. J. biol. Chem. 250, 4007–4021 (1975).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bell, J., Mahadevan, L., Colledge, W. et al. Abelson-transformed fibroblasts contain nuclear phosphotyrosyl-proteins which preferentially bind to murine DNA. Nature 325, 552–554 (1987). https://doi.org/10.1038/325552a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/325552a0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.