Abstract
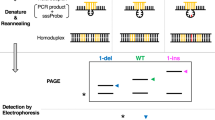
Certain single base substitutions causing genetic diseases1–4 or resulting in polymorphisms linked to mutant alleles5–7, alter a restriction enzyme cleavage site and can therefore be detected in total genomic DNA using DNA blots. Many base substitutions do not lead to an altered restriction site, but these can be detected using synthetic oligonucleotides as hybridization probes if the DNA sequence surrounding the base substitution is known8,9. In the case of β-thalassaemia, where 22 different single base mutations have been identified10, a large number of probes would be required for diagnosis. An approach which was used to detect mutations in viral DNA involves the S1 nuclease treatment of heteroduplexes formed between wild-type and mutant DNA11. Although certain single base mismatches are cleaved by S1 nuclease (ref. 11 and T. Shenk, personal communication), many other mismatches examined by this procedure are not cleaved (B. Seed, personal communication; R.M.M., unpublished data). Heteroduplexes between mutant and wild-type subgenomic fragments of double-stranded reovirus RNA migrate slower than the corresponding homoduplexes in polyacrylamide gels containing 7 M urea12, but it is not known whether this method is applicable to DNA heteroduplexes containing single base mismatches. Here we describe a procedure that involves the electrophoretic separation of DNA heteroduplexes in a well-characterized gel system13. We show that four different human β-thalassaemia alleles with known single base mutations can be detected with as little as 5 µg of total genomic DNA. The method should be useful in the localization and diagnosis of mutations associated with genetic diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Flavell, R. A., Kooter, J. M., De Boer, E., Little, P. F. R. & Williamson, R. Cell 15, 25–41 (1978).
Geever, R. F. et al. Proc. natn. Acad. Sci. U.S.A. 78, 5081–5085 (1981).
Chang, J. C. & Kan, Y. W. New Engl. J. Med. 307, 30–32 (1982).
Orkin, S. H., Little, P. F. R., Kazazian, H. H. & Boehm, C. D. New Engl. J. Med. 307, 32–36 (1982).
Kan, Y. W. & Dozy, A. M. Proc. natn. Acad. Sci. U.S.A. 75, 5631–5635 (1978).
Gusella, J. F. et al. Nature 306, 234–238 (1983).
Orkin, S. H. et al. Nature 296, 627–631 (1982).
Kidd, V. J., Wallace, R. B., Itakura, K. & Woo, S. L. C. Nature 304, 230–234 (1983).
Pirastu, M. et al. New Engl. J. Med. 309, 284–287 (1983).
Orkin, S. H. & Kazazian, H. H. A. Rev. Genet. (in the press).
Shenk, T. E., Rhodes, C., Rigby, P. W. J. & Berg, P. Proc. natn. Acad. Sci. U.S.A. 72, 989–993 (1975).
Ito, Y. & Joklik, W. K. Virology 50, 202–208 (1972).
Fischer, S. G. & Lerman, L. S. Proc. natn. Acad. Sci. U.S.A. 80, 1579–1583 (1983).
Lerman, L. S., Fischer, S. G., Hurley, I., Silverstein, K. & Lumelsky, N. A. Rev. Biophys. Bioengng 13, 399–433 (1984).
Treisman, R., Orkin, S. H. & Maniatis, T. Nature 302, 591–596 (1983).
Atweh, G. F., Hsu, H. & Forget, B. G. Blood 64 (Suppl. 1) (in the press).
Treisman, R., Proudfoot, N. J., Shander, M. & Maniatis, T. Cell 29, 903–911 (1982).
Solomon, E. & Bodmer, W. F. Lancet i, 923 (1979).
Botstein, D., White, R., Skolnick, M. & Davis, R. Am. J. hum. Genet. 32, 314–331 (1980).
Westaway, D. & Williamson, R. Nucleic Acids Res. 9, 1777–1788 (1981).
Spritz, R. A. et al. Proc. natn. Acad. Sci. U.S.A. 78, 2455–2459 (1981).
Lawn, R. M., Fritsch, E. F., Parker, R. C., Blake, G. & Maniatis, T. Cell 15, 1157–1174 (1978).
Levinson, A., Silver, D. & Seed, B. J. molec. appl. Genet. (in the press).
Sanger, F., Coulson, A. R., Barrell, B. G., Smith, A. J. H. & Roe, B. A. J. molec. Biol. 143, 161–178 (1980).
Favalaro, J., Treisman, R. & Kamen, R. Meth. Enzym. 65, 718–749 (1980).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Myers, R., Lumelsky, N., Lerman, L. et al. Detection of single base substitutions in total genomic DNA. Nature 313, 495–498 (1985). https://doi.org/10.1038/313495a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/313495a0
This article is cited by
-
Development of SNP-based CAPS and dCAPS markers in eight different genes involved in starch biosynthesis in rice
Molecular Breeding (2009)
-
Diagnosis of families with familial hypercholesterolaemia and/or Apo B‐100 defect by means of DNA analysis of LDL‐receptor gene mutations
Journal of Inherited Metabolic Disease (2007)
-
The evolution of molecular markers — just a matter of fashion?
Nature Reviews Genetics (2004)
-
Mutation detection and typing of polymorphic loci through double-strand conformation analysis
Nature Genetics (1998)
-
Genotyping HIV-1 and HCV Strains by a Combinatorial DNA Melting Assay (COMA)
Molecular Medicine (1998)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.