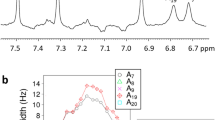
Abstract
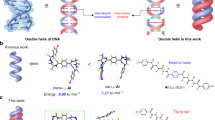
Until recently, all known nucleic acid duplex helices were thought to be conformationally related to A DNA or B DNA, these right-handed helices being characterized by C(3′) endo or C(2′) endo pucker for the respective sugar residues and by an antiglycosidic bond1. This situation has since drastically changed, for X-ray studies on crystals of alternating d(CpG) DNA fragments have demonstrated the existence of a left-handed double helix, termed Z DNA (ref. 2). In this novel double helix the base-pairing is of Watson–Crick type, but the cytidine and guanosine residues have the anti and syn conformations, respectively, and thus the repeating unit is two base pairs. Such a left-handed helix can also account for the X-ray diffraction pattern of alternating poly(dG-dC) ∣ poly(dG-dC) fibres3. However, Pohl and Jovin have shown that in solution poly(dG-dC) ∣ poly(dG-dC) undergoes a salt-induced cooperative transition4. It is very likely that the left-handed double helix is relevant to the high-salt conformation of poly(dG-dC) ∣ poly(dG-dC) and that the low-salt conformation of poly(dG-dC)∣ poly(dG-dC) belongs to the B-DNA family3,5. The techniques previously used to study the salt-induced transition of poly (dG-dC) ∣ poly (dG-dC) in solution—circular dichroism (CD), absorption spectroscopy4 and nuclear magnetic resonance5—give essentially a static picture of the conformation of the molecule. Such information, although fundamental, is incomplete because double-helical nucleic acids in solution are known to be subjected to thermal fluctuations resulting in transient conformations with opened base pairs6–9. A particularly useful probe of the dynamic aspects of nucleic acid structure is the tritium exchange technique largely developed by Englander and colleagues10. We have now used this technique to study the dynamic structure of poly(dG-dC) · poly (dG-dC) and have found that this varies depending on whether high or low salt conditions are used.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Crick, F. H. C., Wang, J. C. & Bauer, W. R. J. molec. Biol. 129, 449–461 (1979).
Wang, A. H. J. et al. Nature 282, 680–686 (1979).
Arnott, S., Chandrasekaran, R., Birdsall, D. L., Leslie, A. G. W. & Ratliflf, R. L. Nature 283, 743–745 (1980).
Pohl, F. M. & Jovin, T. M. J. molec. Biol. 67, 375–396 (1972).
Patel, D.J., Canuel, L.L. & Pohl, F.M. Proc. natn. Acad. Sci. U.S.A. 76, 2508–2511 (1979).
Teitelbaum, H. & Englander, S. W. J. molec. Biol. 92, 55–78 (1975).
Teitelbaum, H. & Englander, S. W. J. molec. Biol. 92, 79–92 (1975).
McConnel, B. & Von Hippel, P. H. J. molec. Biol. 50, 297–316 (1970).
McConnel, B. & Von Hippel, P. H. J. molec. Biol. 50, 317–332 (1970).
Englander, S. W. & Englander, J. J. Meth. Enzym. 26C, 406–413 (1972).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ramstein, J., Leng, M. Salt-dependent dynamic structure of poly(dG-dC) · poly(dG-dC). Nature 288, 413–414 (1980). https://doi.org/10.1038/288413a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/288413a0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.