Abstract
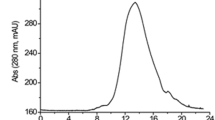
THE recent development in this laboratory of methods for producing acid proteinase of Aspergillus saitoi (EC, 3.4.4.17, aspergillopeptidase A) from culture filtrate has made available sufficient material for a correlative study of its enzymatic properties1,2. The optimal pH for milk casein digestion is in the pH range of 2.5–3.0 and the proteinase is fairly stable over the pH range of 2.5–6.0 (ref. 3). Aspergillopeptidase A is capable of activating trypsinogen and chymotrypsinogen A at pH 4.5 (ref. 4). The further purification method for separating acid proteinase has been improved by chromatographic procedures with the use of ion exchange materials, ‘Duolite CS-101’, DEAE-cellulose and ‘SE-Sephadex’5. The purified aspergillopeptidase A appears to be homogeneous on free boundary electrophoresis over a pH range of 2.0–10.0 and on ultracentrifugation at pH 4.1. The iso-electric point has been found to be at pH 3.65 in Sørensen's citrate buffer. In the investigation reported here, the molecular weight of aspergillopeptidase A was found to be 34,900 from sedimentation and viscosity, according to Scheraga–Mandel-kern's formula6,7, and was also determined to be 34,200 according to Yphantis's treatment8.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Yoshida, F., J. Agric. Chem. Soc. Japan, 28, 66 (1954). Bull. Agric. Chem. Soc. Japan, 20, 252 (1956).
Ichishima, E., and Yoshida, F., Agric. Biol. Chem. Japan, 26, 547, 554 (1962); 27, 302, 310 (1963).
Yoshida, F., and Nagasawa, M., Bull. Agric. Chem. Soc. Japan, 20, 257 (1956).
Gabeloteau, C., and Desnuelle, P., Biochim. Biophys. Acta, 42, 230 (1960).
Ichishima, E., and Yoshida, F., Biochim. Biophys. Acta, 99, 360 (1965).
Scheraga, H. A., and Mandelkern, L., J. Amer. Chem. Soc., 75, 179 (1953).
Schachman, H. K., in Methods in Enzymology, edit. by Colowick, S. P., and Kaplan, N. O., 32 (Academic Press, New York, 1957).
Yphantis, D. A., Ann. N.Y. Acad. Sci., 88, 586 (1960).
Schachman, H. K., Ultracentrifugation in Biochemistry (Academic Press, New York, 1959).
Ichishima, E., and Yoshida, F., Biochim. Biophys. Acta (in the press).
Meekin, T. J., Groves, M. L., and Hipp, N. J., J. Amer. Chem. Soc., 71, 3298 (1949).
Cohn, E. J., and Edsall, J. T., Proteins, Amino Acids and Peptides, 370 (Reinhold, New York, 1943).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
ICHISHIMA, E., YOSHIDA, F. Molecular Weight of Acid Proteinase of Aspergillus saitoi. Nature 207, 525–526 (1965). https://doi.org/10.1038/207525a0
Published:
Issue Date:
DOI: https://doi.org/10.1038/207525a0
This article is cited by
-
Tryptophan Residue in Aspergillopeptidase A
Nature (1967)
-
Molecular weight of cathepsins from different animal organs
Die Naturwissenschaften (1966)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.