Abstract
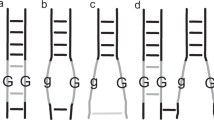
STRICKLAND1 has applied to tetrad data for Aspergillus nidulans the method I have suggested2 for estimating the magnitude of chromatid interference between cross-overs when loosely linked genes are used. The principle underlying this method is as follows : Consider three linked loci X, Y and Z spaced along a chromosome in that order. By means of tetrad analysis one can distinguish a number of tetrad genotypes. Certain of these, called (a), (b) and (c), respectively, are most simply interpreted as resulting from the following cross-overs at meiosis : (a) One cross-over between X and Y and another between Y and Z involving the same chromatids, that is, a two-strand double cross-over. (b) One cross-over between X and Y and another between Y and Z, with a three-strand relationship. (c) One cross-over between X and Y and another between Y and Z involving the other two chromatids, that is, a four-strand double cross-over. With closely linked genes, the frequencies of (a), (b) and (c) provide accurate information about the frequencies of two-, three-and four-strand relationships between cross-overs. If the genes are loosely linked, these tetrad genotypes can also arise as a consequence of a three-strand double cross-over between X and Y (or Y and Z) and a third cross-over between Y and Z (or X and Y), but the strand-relationships with the third cross-over are two- or four-strand for genotype (b) and three-strand for genotypes (a) and (c). In other words, the occurrence of these triple cross-over tetrads will hinder the detection of chromatid interference by tending to obscure differences from the 1 : 2 : 1 proportion of genotypes (a), (b) and (c). This is the ratio expected with no chromatid interference. The frequency of these inconvenient triple cross-over tetrads can be estimated from the observed frequency of another tetrad genotype (d), which results from a four-strand double cross-over between X and Y (or Y and Z) and a third cross-over between Y and Z (or X and Y). In this way, allowance can be made for the effects of triple cross-overs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Strickland, W. N., Proc. Roy. Soc., B, 149, 82 (1958).
Whitehouse, H. L. K., C.R. Lab. Carlsberg, 26, 407 (1956).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
WHITEHOUSE, H. Use of Loosely Linked Genes to estimate Chromatid Interference by Tetrad Analysis. Nature 182, 1173–1174 (1958). https://doi.org/10.1038/1821173a0
Issue Date:
DOI: https://doi.org/10.1038/1821173a0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.